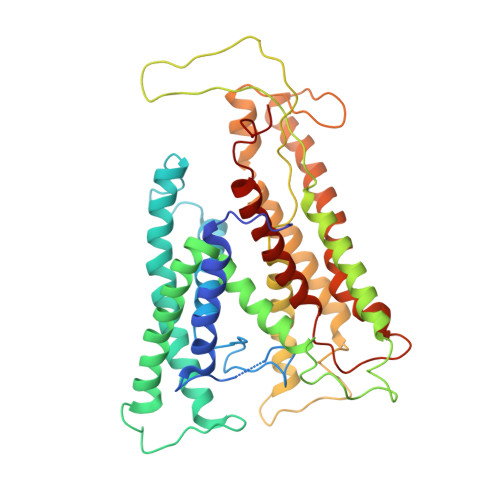
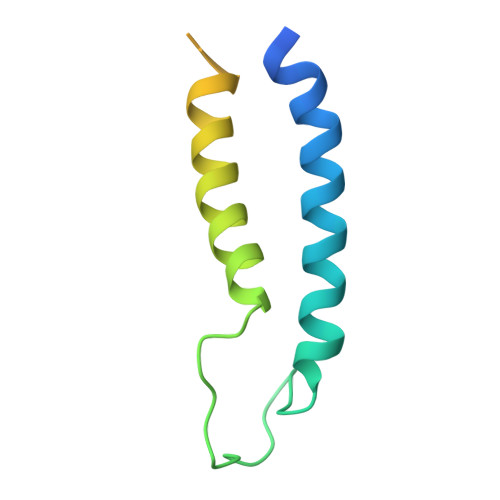
Structure of a complex of the ATPase SecA and the protein-translocation channel.
Zimmer, J., Nam, Y., Rapoport, T.A.(2008) Nature 455: 936-943
- PubMed: 18923516
- DOI: https://doi.org/10.1038/nature07335
- Primary Citation of Related Structures:
3DIN, 3DL8 - PubMed Abstract:
Most proteins are secreted from bacteria by the interaction of the cytoplasmic SecA ATPase with a membrane channel, formed by the heterotrimeric SecY complex. Here we report the crystal structure of SecA bound to the SecY complex, with a maximum resolution of 4.5 ångström (A), obtained for components from Thermotoga maritima. One copy of SecA in an intermediate state of ATP hydrolysis is bound to one molecule of the SecY complex. Both partners undergo important conformational changes on interaction. The polypeptide-cross-linking domain of SecA makes a large conformational change that could capture the translocation substrate in a 'clamp'. Polypeptide movement through the SecY channel could be achieved by the motion of a 'two-helix finger' of SecA inside the cytoplasmic funnel of SecY, and by the coordinated tightening and widening of SecA's clamp above the SecY pore. SecA binding generates a 'window' at the lateral gate of the SecY channel and it displaces the plug domain, preparing the channel for signal sequence binding and channel opening.
- Howard Hughes Medical Institute and Department of Cell Biology, Harvard Medical School, 240 Longwood Avenue, Boston, Massachusetts 02115, USA.
Organizational Affiliation: