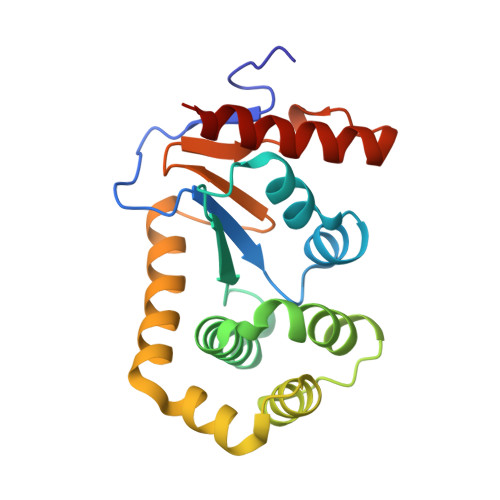
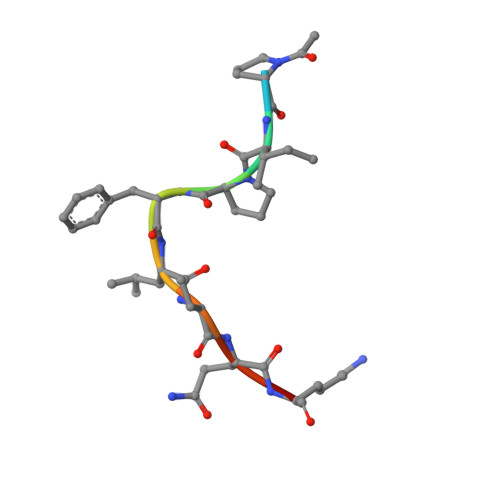
The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
Paxman, J.J., Borg, N.A., Horne, J., Thompson, P.E., Chin, Y., Sharma, P., Simpson, J.S., Wielens, J., Piek, S., Kahler, C.M., Sakellaris, H., Pearce, M., Bottomley, S.P., Rossjohn, J., Scanlon, M.J.(2009) J Biological Chem 284: 17835-17845
- PubMed: 19389711
- DOI: https://doi.org/10.1074/jbc.M109.011502
- Primary Citation of Related Structures:
3DKS - PubMed Abstract:
Oxidative protein folding in Gram-negative bacteria results in the formation of disulfide bonds between pairs of cysteine residues. This is a multistep process in which the dithiol-disulfide oxidoreductase enzyme, DsbA, plays a central role. The structure of DsbA comprises an all helical domain of unknown function and a thioredoxin domain, where active site cysteines shuttle between an oxidized, substrate-bound, reduced form and a DsbB-bound form, where DsbB is a membrane protein that reoxidizes DsbA. Most DsbA enzymes interact with a wide variety of reduced substrates and show little specificity. However, a number of DsbA enzymes have now been identified that have narrow substrate repertoires and appear to interact specifically with a smaller number of substrates. The transient nature of the DsbA-substrate complex has hampered our understanding of the factors that govern the interaction of DsbA enzymes with their substrates. Here we report the crystal structure of a complex between Escherichia coli DsbA and a peptide with a sequence derived from a substrate. The binding site identified in the DsbA-peptide complex was distinct from that observed for DsbB in the DsbA-DsbB complex. The structure revealed details of the DsbA-peptide interaction and suggested a mechanism by which DsbA can simultaneously show broad specificity for substrates yet exhibit specificity for DsbB. This mode of binding was supported by solution nuclear magnetic resonance data as well as functional data, which demonstrated that the substrate specificity of DsbA could be modified via changes at the binding interface identified in the structure of the complex.
- Medicinal Chemistry and Drug Action, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville Campus, 381 Royal Parade, Parkville, Victoria 3052, Australia.
Organizational Affiliation: