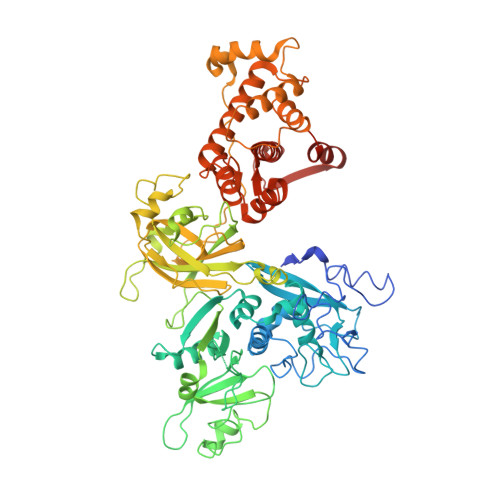
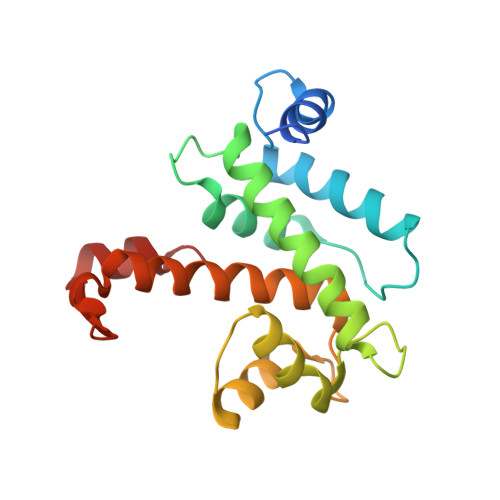
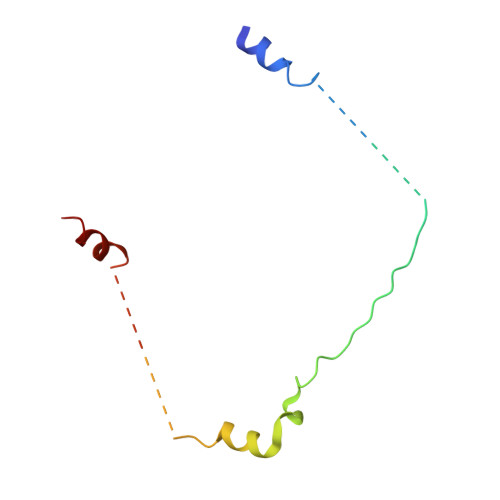
Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Moldoveanu, T., Gehring, K., Green, D.R.(2008) Nature 456: 404-408
- PubMed: 19020622
- DOI: https://doi.org/10.1038/nature07353
- Primary Citation of Related Structures:
3DF0 - PubMed Abstract:
The Ca(2+)-dependent cysteine proteases, calpains, regulate cell migration, cell death, insulin secretion, synaptic function and muscle homeostasis. Their endogenous inhibitor, calpastatin, consists of four inhibitory repeats, each of which neutralizes an activated calpain with exquisite specificity and potency. Despite the physiological importance of this interaction, the structural basis of calpain inhibition by calpastatin is unknown. Here we report the 3.0 A structure of Ca(2+)-bound m-calpain in complex with the first calpastatin repeat, both from rat, revealing the mechanism of exclusive specificity. The structure highlights the complexity of calpain activation by Ca(2+), illustrating key residues in a peripheral domain that serve to stabilize the protease core on Ca(2+) binding. Fully activated calpain binds ten Ca(2+) atoms, resulting in several conformational changes allowing recognition by calpastatin. Calpain inhibition is mediated by the intimate contact with three critical regions of calpastatin. Two regions target the penta-EF-hand domains of calpain and the third occupies the substrate-binding cleft, projecting a loop around the active site thiol to evade proteolysis.
- Department of Immunology, St Jude Children's Research Hospital, 332 N Lauderdale, Memphis, Tennessee 38105, USA.
Organizational Affiliation: