Crystal structure of cAMP receptor protein from M. tuberculosis in the unliganded form
Gallagher, D.T., Robinson, H., Smith, N., Reddy, P.T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TRANSCRIPTIONAL REGULATORY PROTEIN | 227 | Mycobacterium tuberculosis | Mutation(s): 0 Gene Names: MT3777, Rv3676 | 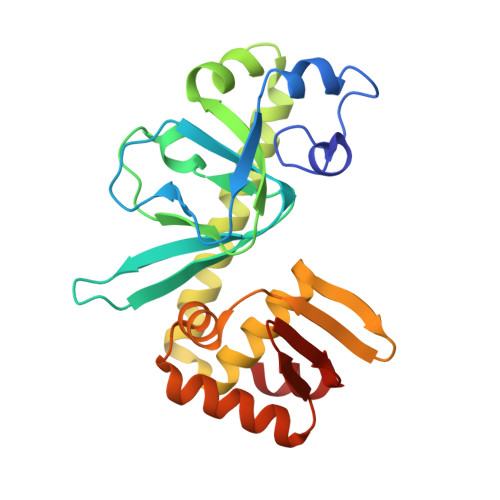 | |
UniProt | |||||
Find proteins for P9WMH3 (Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)) Explore P9WMH3 Go to UniProtKB: P9WMH3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P9WMH3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL Query on CL | C [auth A], D [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 52.796 | α = 90 |
| b = 83.803 | β = 90 |
| c = 98.389 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| RESOLVE | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-3000 | data collection |
| PHENIX | phasing |