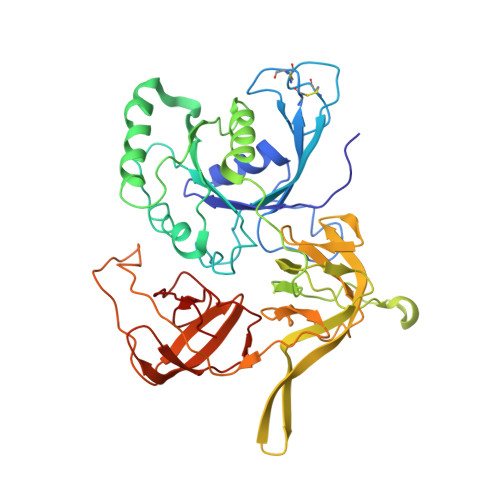
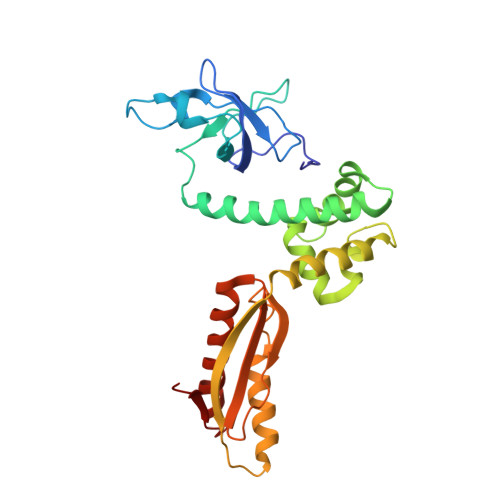
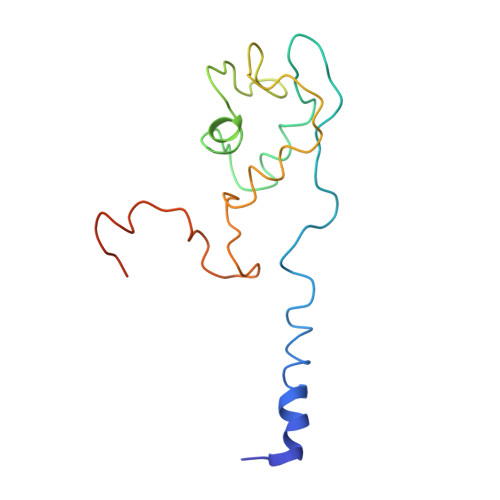
Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the alpha- and beta-subunits.
Stolboushkina, E., Nikonov, S., Nikulin, A., Blasi, U., Manstein, D.J., Fedorov, R., Garber, M., Nikonov, O.(2008) J Mol Biology 382: 680-691
- PubMed: 18675278
- DOI: https://doi.org/10.1016/j.jmb.2008.07.039
- Primary Citation of Related Structures:
3CW2 - PubMed Abstract:
In Eukarya and Archaea, translation initiation factor 2 (eIF2/aIF2), which contains three subunits (alpha, beta, and gamma), is pivotal for binding of charged initiator tRNA to the small ribosomal subunit. The crystal structure of the full-sized heterotrimeric aIF2 from Sulfolobus solfataricus in the nucleotide-free form has been determined at 2.8-A resolution. Superposition of four molecules in the asymmetric unit of the crystal and the comparison of the obtained structures with the known structures of the aIF2alphagamma and aIF2betagamma heterodimers revealed high conformational flexibility in the alpha- and beta-subunits. In fact, the full-sized aIF2 consists of a rigid central part, formed by the gamma-subunit, domain 3 of the alpha-subunit, and the N-terminal alpha-helix of the beta-subunit, and two mobile "wings," formed by domains 1 and 2 of the alpha-subunit, the central part, and the zinc-binding domain of the beta-subunit. High structural flexibility of the wings is probably required for interaction of aIF2 with the small ribosomal subunit. Comparative analysis of all known structures of the gamma-subunit alone and within the heterodimers and heterotrimers in nucleotide-bound and nucleotide-free states shows that the conformations of switch 1 and switch 2 do not correlate with the assembly or nucleotide states of the protein.
- Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russian Federation.
Organizational Affiliation: