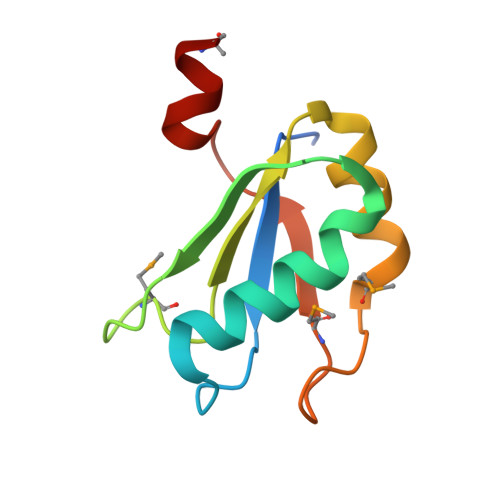
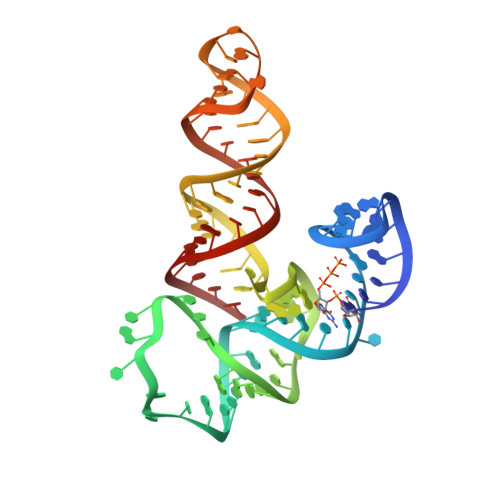
Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Xiao, H., Murakami, H., Suga, H., Ferre-D'Amare, A.R.(2008) Nature 454: 358-361
- PubMed: 18548004
- DOI: https://doi.org/10.1038/nature07033
- Primary Citation of Related Structures:
3CUL, 3CUN - PubMed Abstract:
In modern organisms, protein enzymes are solely responsible for the aminoacylation of transfer RNA. However, the evolution of protein synthesis in the RNA world required RNAs capable of catalysing this reaction. Ribozymes that aminoacylate RNA by using activated amino acids have been discovered through selection in vitro. Flexizyme is a 45-nucleotide ribozyme capable of charging tRNA in trans with various activated l-phenylalanine derivatives. In addition to a more than 10(5) rate enhancement and more than 10(4)-fold discrimination against some non-cognate amino acids, this ribozyme achieves good regioselectivity: of all the hydroxyl groups of a tRNA, it exclusively aminoacylates the terminal 3'-OH. Here we report the 2.8-A resolution structure of flexizyme fused to a substrate RNA. Together with randomization of ribozyme core residues and reselection, this structure shows that very few nucleotides are needed for the aminoacylation of specific tRNAs. Although it primarily recognizes tRNA through base-pairing with the CCA terminus of the tRNA molecule, flexizyme makes numerous local interactions to position the acceptor end of tRNA precisely. A comparison of two crystallographically independent flexizyme conformations, only one of which appears capable of binding activated phenylalanine, suggests that this ribozyme may achieve enhanced specificity by coupling active-site folding to tRNA docking. Such a mechanism would be reminiscent of the mutually induced fit of tRNA and protein employed by some aminoacyl-tRNA synthetases to increase specificity.
- Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, Washington 98109-1024, USA.
Organizational Affiliation: