Structural analysis of the Saf pilus by electron microscopy and image processing.
Salih, O., Remaut, H., Waksman, G., Orlova, E.V.(2008) J Mol Biology 379: 174-187
- PubMed: 18448124
- DOI: https://doi.org/10.1016/j.jmb.2008.03.056
- Primary Citation of Related Structures:
3CRE, 3CRF - PubMed Abstract:
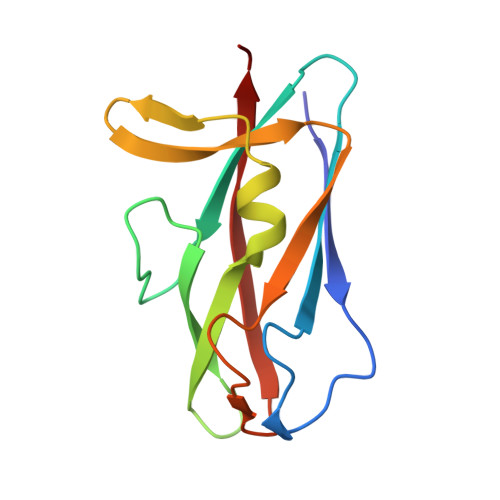
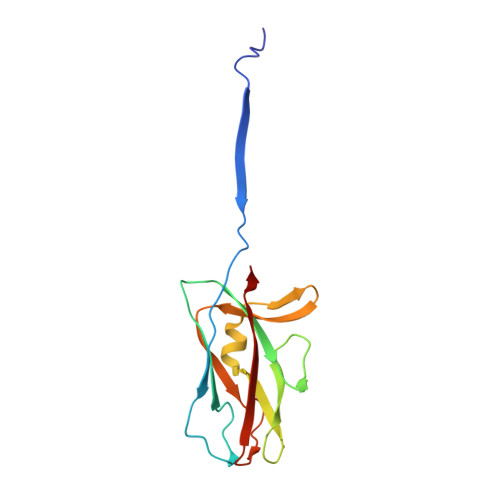
Bacterial pili are important virulence factors involved in host cell attachment and/or biofilm formation, key steps in establishing and maintaining successful infection. Here we studied Salmonella atypical fimbriae (or Saf pili), formed by the conserved chaperone/usher pathway. In contrast to the well-established quaternary structure of typical/FGS-chaperone assembled, rod-shaped, chaperone/usher pili, little is known about the supramolecular organisation in atypical/FGL-chaperone assembled fimbriae. In our study, we have used negative stain electron microscopy and single-particle image analysis to determine the three-dimensional structure of the Salmonella typhimurium Saf pilus. Our results show atypical/FGL-chaperone assembled fimbriae are composed of highly flexible linear multi-subunit fibres that are formed by globular subunits connected to each other by short links giving a "beads on a string"-like appearance. Quantitative fitting of the atomic structure of the SafA pilus subunit into the electron density maps, in combination with linker modelling and energy minimisation, has enabled analysis of subunit arrangement and intersubunit interactions in the Saf pilus. Short intersubunit linker regions provide the molecular basis for flexibility of the Saf pilus by acting as molecular hinges allowing a large range of movement between consecutive subunits in the fibre.
- Institute of Structural Molecular Biology at UCL/Birkbeck, London WC1E 7HX, UK.
Organizational Affiliation: