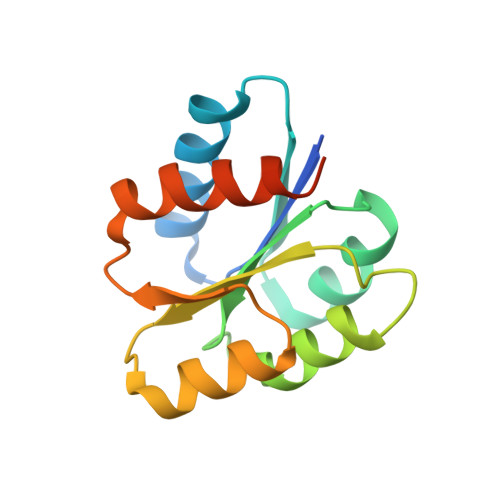
Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
Patskovsky, Y., Romero, R., Freeman, J., Hu, S., Groshong, C., Wasserman, S.R., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.