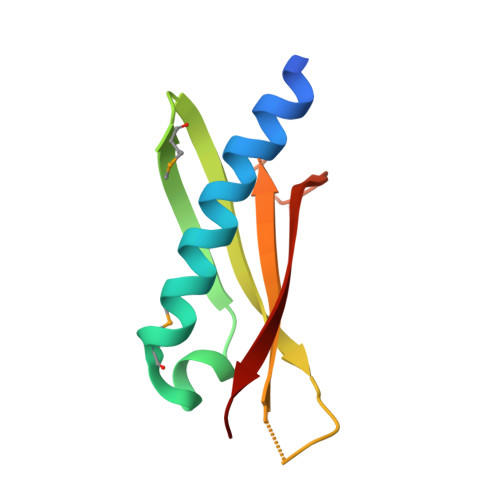
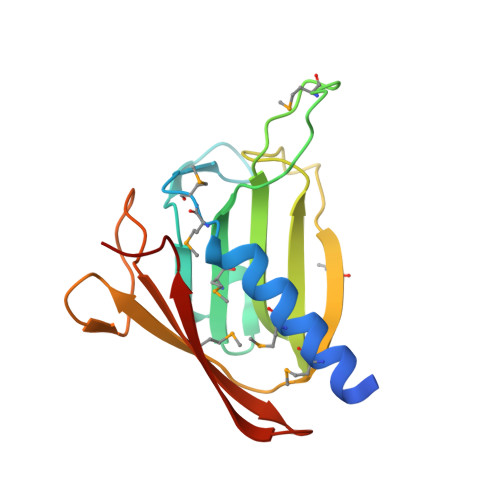
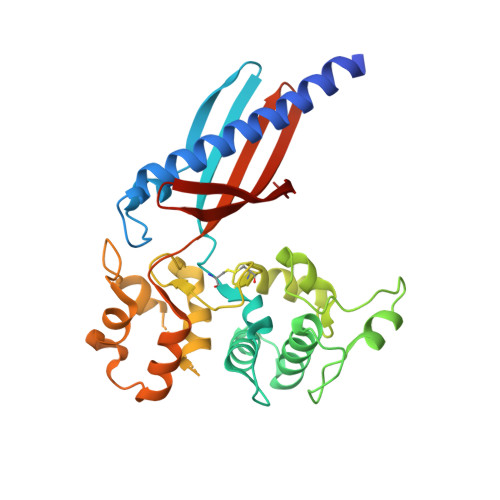
Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system.
Korotkov, K.V., Hol, W.G.(2008) Nat Struct Mol Biol 15: 462-468
- PubMed: 18438417
- DOI: https://doi.org/10.1038/nsmb.1426
- Primary Citation of Related Structures:
3CI0 - PubMed Abstract:
Gram-negative bacteria translocate various proteins including virulence factors across their outer membrane via type 2 secretion systems (T2SSs). T2SSs are thought to contain a pseudopilus, a subcomplex formed by one major and several minor pseudopilins. We report the crystal structure of the complex formed by three minor pseudopilins from enterotoxigenic Escherichia coli. The GspK-GspI-GspJ complex has quasihelical characteristics and an architecture consistent with a localization at the pseudopilus tip. The alpha-domain of GspK has a previously unobserved fold with an unexpected dinuclear metal binding site. The area surrounding its disulfide bridge is conserved and might interact with other T2SS components or with secreted proteins.
- Department of Biochemistry, Biomolecular Structure Center, University of Washington, Box 357742, Seattle, Washington 98195, USA.
Organizational Affiliation: