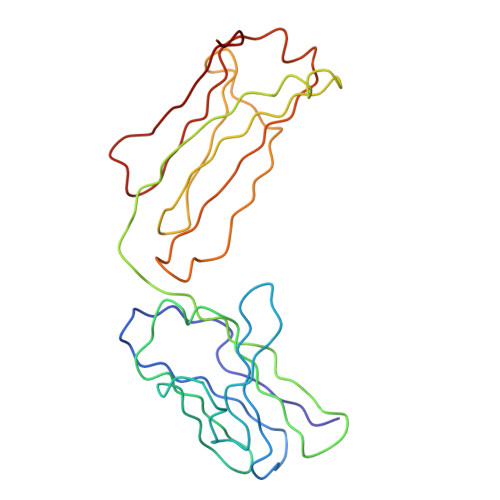
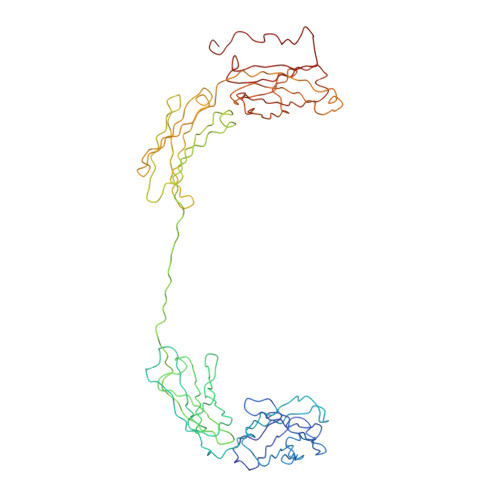
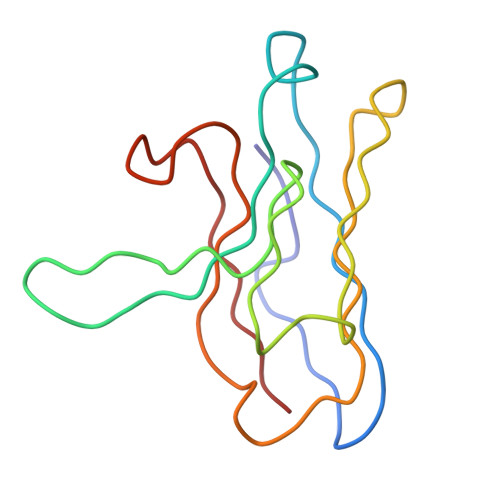
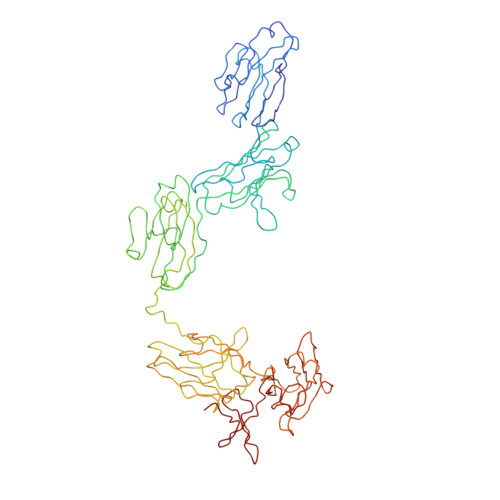
Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity.
Bonner, A., Almogren, A., Furtado, P.B., Kerr, M.A., Perkins, S.J.(2009) Mucosal Immunol 2: 74-84
- PubMed: 19079336
- DOI: https://doi.org/10.1038/mi.2008.68
- Primary Citation of Related Structures:
3CHN - PubMed Abstract:
Secretory immunoglobulin A (SIgA) is the most prevalent antibody in the human body and a first line of defense in mucosal immunity. We located secretory component (SC) relative to dimeric IgA1 (dIgA1) within the SIgA1 structure using the constrained modeling of solution scattering and analytical ultracentrifugation data. The extended solution structure of dIgA1 is largely preserved within SIgA1. From conformational searches of SC locations, the best-fit SC models within SIgA1 show that SC is extended along the outermost convex edge of the Fc dimer in dIgA1. The topology of our SIgA1 structure reveals that it is able to bind to one FcalphaRI receptor molecule. SC binding to the Fc dimer confers protection to SIgA1 by the masking of proteolytically susceptible surface sites from bacterial proteases in the harsh environment of the mucosa. The models support a "zipper-like" unfolding of SC upon dIgA1 in the formation and transportation of SIgA1 into the mucosa.
- Division of Biosciences, Institute of Structural and Molecular Biology, University College London, London, UK.
Organizational Affiliation: