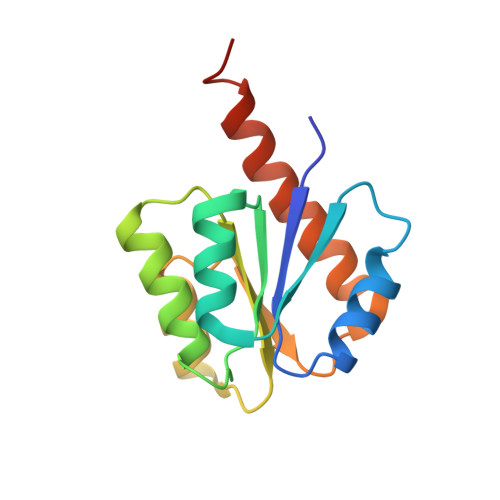
Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
Patskovsky, Y., Ramagopal, U.A., Fong, R., Freeman, J., Iizuka, M., Groshong, C., Smith, D., Wasserman, S.R., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.