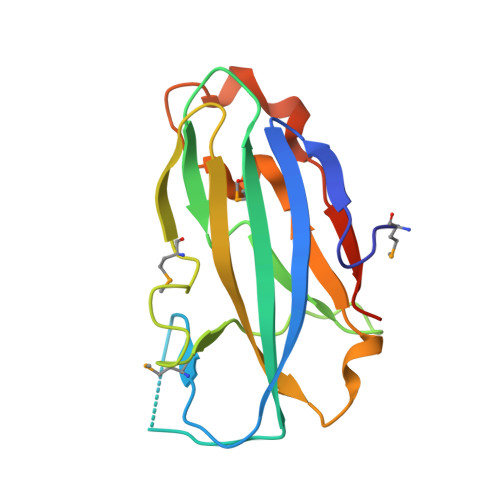
Crystal structure of the yjhA protein from Bacillus subtilis.
Vorobiev, S.M., Chen, Y., Kuzin, A.P., Seetharaman, J., Forouhar, F., Zhao, L., Mao, L., Maglaqui, M., Xiao, R., Liu, J., Swapna, G., Huang, J.Y., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.