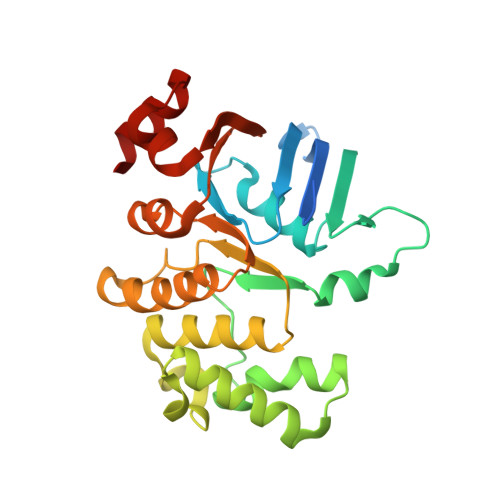
Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
Thaben, P.F., Eckey, V., Scheffel, F., Allings, C., Behlke, J., Saenger, W., Schneider, E., Vahedi-Faridi, A.To be published.