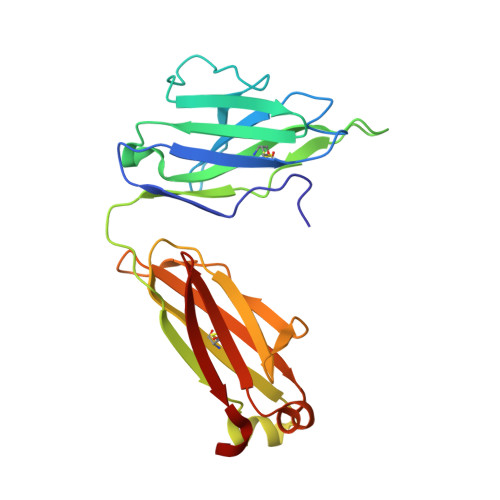
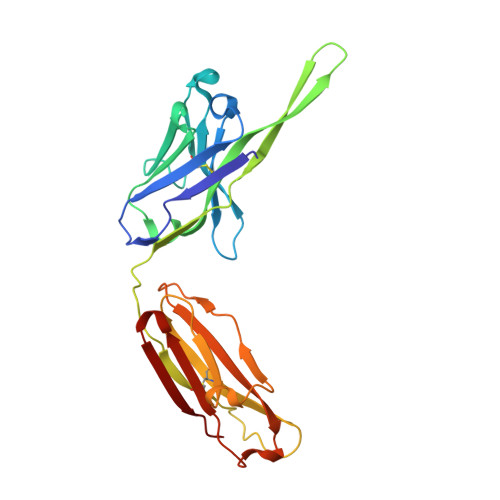
Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set
Dhillon, A.K., Stanfield, R.L., Gorny, M.K., Williams, C., Zolla-Pazner, S., Wilson, I.A.(2008) Acta Crystallogr D Biol Crystallogr 64: 792-802
- PubMed: 18566514
- DOI: https://doi.org/10.1107/S0907444908013978
- Primary Citation of Related Structures:
3C2A - PubMed Abstract:
Although antibodies against the third variable loop (V3) of the HIV-1 viral envelope glycoprotein are among the first neutralizing antibodies to be detected in infected individuals, they are normally restricted in their specificity. X-ray crystallographic studies of V3-specific antibodies have contributed to a more thorough understanding of recognition of this epitope and of conserved features in the V3 loop that could potentially aid in the design of a multi-component vaccine. The human antibody 447-52D exhibits relatively broad neutralization of primary viral isolates compared with other V3-loop antibodies. A crystal structure of Fab 447-52D in complex with a V3 peptide (UG1033) was determined at 2.1 angstroms resolution. The structure was determined using an epitaxially twinned data set and in-house programs to detect and remove overlapping reflections. Although the processed data have lower than desired completeness and slightly higher than normal R values for the resolution, good-quality electron-density maps were obtained that enabled structure determination. The structure revealed an extended CDR H3 loop that forms a beta-sheet with the peptide, with the predominant contacts being main-chain hydrogen bonds. The V3 peptide and Fab show high structural homology with the previously reported structures of other Fab 447-52D complexes, reinforcing the idea that the V3 loop may adopt a small set of conserved structures, particularly around the crown of the beta-hairpin.
- Department of Immunology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, USA.
Organizational Affiliation: