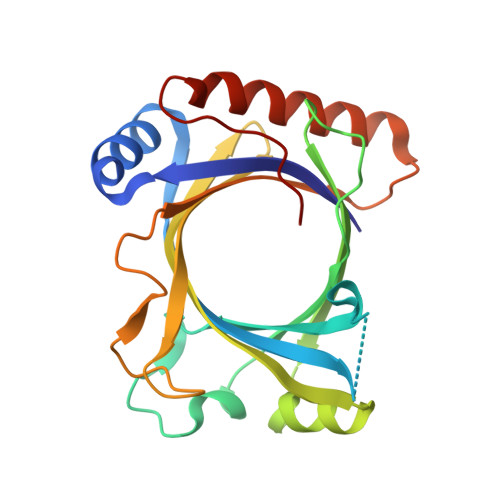
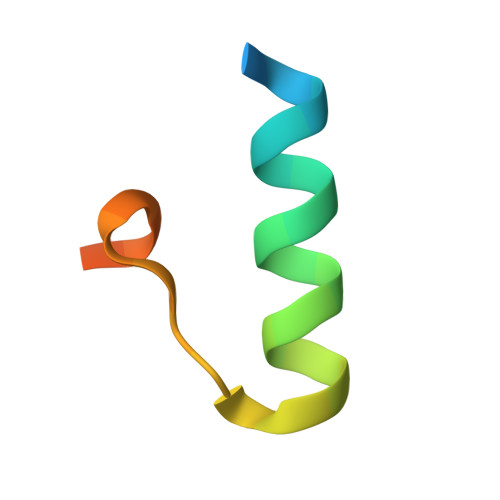
Structure-system correlation identifies a gene regulatory Mediator submodule
Lariviere, L., Seizl, M., van Wageningen, S., Rother, S., van de Pasch, L., Feldmann, H., Strasser, K., Hahn, S., Holstege, F.C.P., Cramer, P.(2008) Genes Dev 22: 872-877
- PubMed: 18381891
- DOI: https://doi.org/10.1101/gad.465108
- Primary Citation of Related Structures:
3C0T - PubMed Abstract:
A combination of crystallography, biochemistry, and gene expression analysis identifies the coactivator subcomplex Med8C/18/20 as a functionally distinct submodule of the Mediator head module. Med8C forms a conserved alpha-helix that tethers Med18/20 to the Mediator. Deletion of Med8C in vivo results in dissociation of Med18/20 from Mediator and in loss of transcription activity of extracts. Deletion of med8C, med18, or med20 causes similar changes in the yeast transcriptome, establishing Med8C/18/20 as a predominantly positive, gene-specific submodule required for low transcription levels of nonactivated genes, including conjugation genes. The presented structure-based system perturbation is superior to gene deletion analysis of gene regulation.
- Gene Center Munich and Center for Integrated Protein Science CIPSM, Department of Chemistry and Biochemistry, Ludwig-Maximilians-Universität München, 81377 Munich, Germany.
Organizational Affiliation: