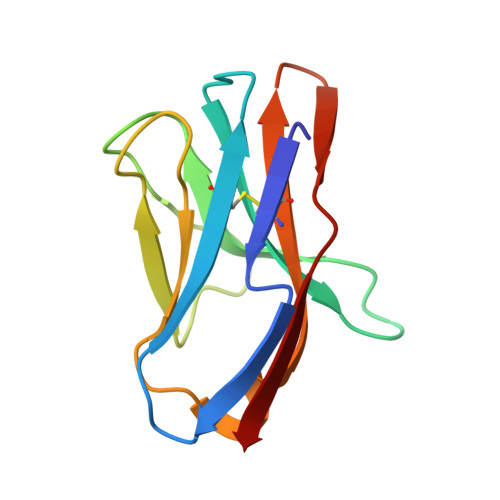
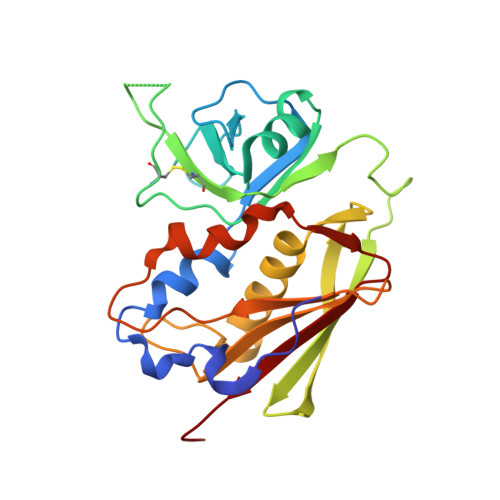
Manipulating the coupled folding and binding process drives affinity maturation in a protein-protein complex
Cho, S., Swaminathan, C.P., Kerzic, M.C., Guan, R., Yang, J., Kieke, M.C., Andersen, P.S., Krantz, D.M., Mariuzza, R.A., Eric, S.J.To be published.