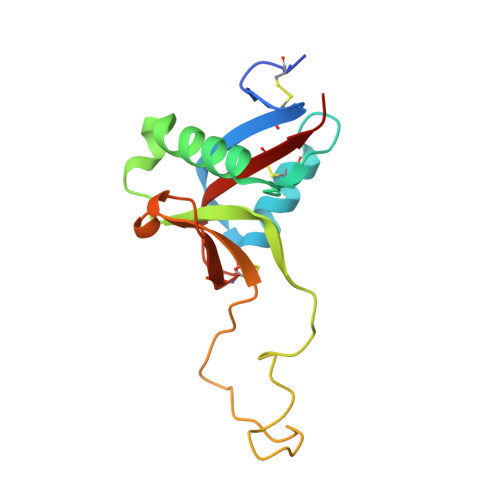
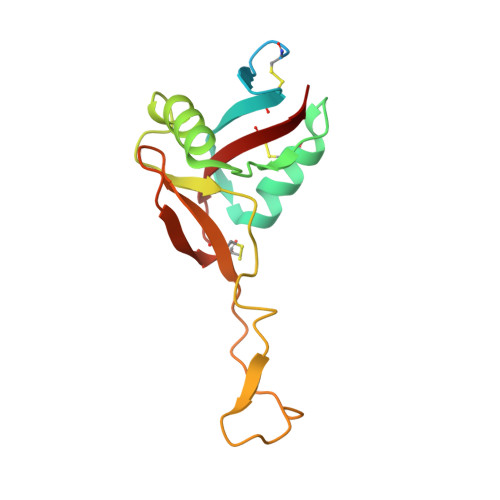
The crystal structure of the platelet activator aggretin reveals a novel (alphabeta)2 dimeric structure.
Hooley, E., Papagrigoriou, E., Navdaev, A., Pandey, A.V., Clemetson, J.M., Clemetson, K.J., Emsley, J.(2008) Biochemistry 47: 7831-7837
- PubMed: 18597489
- DOI: https://doi.org/10.1021/bi800528t
- Primary Citation of Related Structures:
3BX4 - PubMed Abstract:
Aggretin is a C-type lectin purified from Calloselasma rhodostoma snake venom. It is a potent activator of platelets, resulting in a collagen-like response by binding and clustering platelet receptor CLEC-2. We present here the crystal structure of aggretin at 1.7 A which reveals a unique tetrameric quaternary structure. The two alphabeta heterodimers are arranged through 2-fold rotational symmetry, resulting in an antiparallel side-by-side arrangement. Aggretin thus presents two ligand binding sites on one surface and can therefore cluster ligands in a manner reminiscent of convulxin and flavocetin. To examine the molecular basis of the interaction with CLEC-2, we used a molecular modeling approach of docking the aggretin alphabeta structure with the CLEC-2 N-terminal domain (CLEC-2N). This model positions the CLEC-2N structure face down in the "saddle"-shaped binding site which lies between the aggretin alpha and beta lectin-like domains. A 2-fold rotation of this complex to generate the aggretin tetramer reveals dimer contacts for CLEC-2N which bring the N- and C-termini into the proximity of each other, and a series of contacts involving two interlocking beta-strands close to the N-terminus are described. A comparison with homologous lectin-like domains from the immunoreceptor family reveals a similar but not identical dimerization mode, suggesting this structure may represent the clustered form of CLEC-2 capable of signaling across the platelet membrane.
- Centre for Biomolecular Sciences, School of Pharmacy, University of Nottingham, UK.
Organizational Affiliation: