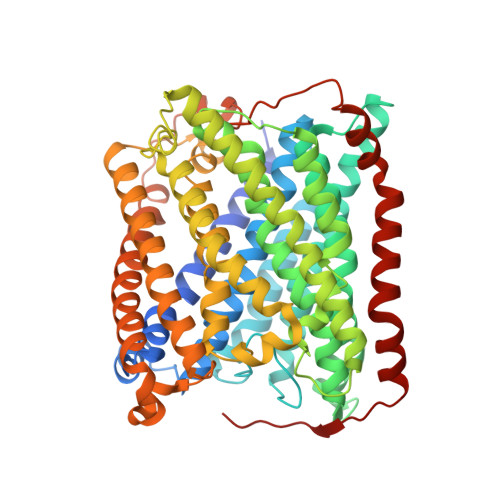
Crystallographic Studies of Xe and Kr Binding within the Large Internal Cavity of Cytochrome ba3 from Thermus thermophilus: Structural Analysis and Role of Oxygen Transport Channels in the Heme-Cu Oxidases.
Luna, V.M., Chen, Y., Fee, J.A., Stout, C.D.(2008) Biochemistry 47: 4657-4665
- PubMed: 18376849
- DOI: https://doi.org/10.1021/bi800045y
- Primary Citation of Related Structures:
3BVD - PubMed Abstract:
Cytochrome ba3 is a cytochrome c oxidase from the plasma membrane of Thermus thermophilus and is the preferred terminal enzyme of cellular respiration at low dioxygen tensions. Using cytochrome ba 3 crystals pressurized at varying conditions under Xe or Kr gas, and X-ray data for six crystals, we identify the relative affinities of Xe and Kr atoms for as many as seven distinct binding sites. These sites track a continuous, Y-shaped channel, 18-20 A in length, lined by hydrophobic residues, which leads from the surface of the protein where two entrance holes, representing the top of the Y, connect the bilayer to the a3-CuB center at the base of the Y. Considering the increased affinity of O2 for hydrophobic environments, the hydrophobic nature of the channel, its orientation within the bilayer, its connection to the active site, its uniform diameter, its virtually complete occupation by Xe, and its isomorphous presence in the native enzyme, we infer that the channel is a diffusion pathway for O2 into the dinuclear center of cytochrome ba3. These observations provide a basis for analyzing similar channels in other oxidases of known structure, and these structures are discussed in terms of mechanisms of O2 transport in biological systems, details of CO binding to and egress from the dinuclear center, the bifurcation of the oxygen-in and water-out pathways, and the possible role of the oxygen channel in aerobic thermophily.
- Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: