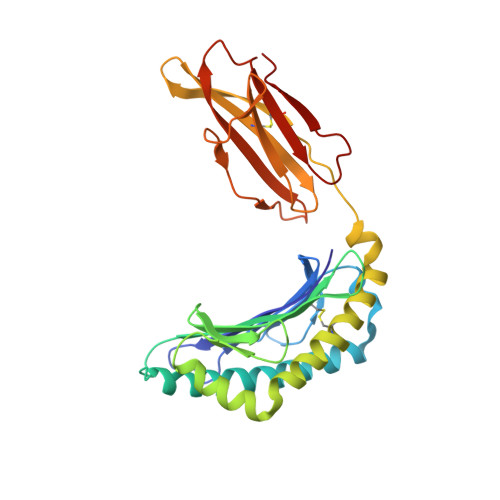
Epitope-specific TCRbeta repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides
La Gruta, N.L., Thomas, P.G., Webb, A.I., Dunstone, M.A., Cukalac, T., Doherty, P.C., Purcell, A.W., Rossjohn, J., Turner, S.J.(2008) Proc Natl Acad Sci U S A 105: 2034-2039
- PubMed: 18238896
- DOI: https://doi.org/10.1073/pnas.0711682102
- Primary Citation of Related Structures:
3BUY - PubMed Abstract:
TCR repertoire diversity has been convincingly shown to facilitate responsiveness of CD8+ T cell populations to mutant virus peptides, thereby safeguarding against viral escape. However, the impact of repertoire diversity on the functionality of the CD8+ T cell response to cognate peptide-MHC class I complex (pMHC) recognition remains unclear. Here, we have compared TCRbeta chain repertoires of three influenza A epitope-specific CD8+ T cell responses in C57BL/6 (B6) mice: D(b)NP(366-374), D(b)PA(224-233), and a recently described epitope derived from the +1 reading frame of the influenza viral polymerase B subunit (residues 62-70) (D(b)PB1-F2(62)). Corresponding to the relative antigenicity of the respective pMHCs, and irrespective of the location of prominent residues, the D(b)PA(224)- and D(b)PB1-F2(62)-specific repertoires were similarly diverse, whereas the D(b)NP(366) population was substantially narrower. Importantly, parallel analysis of response magnitude, cytotoxicity, TCR avidity, and cytokine production for the three epitope-specific responses revealed no obvious functional advantage conferred by increased T cell repertoire diversity. Thus, whereas a diverse repertoire may be important for recognition of epitope variants, its effect on the response to cognate pMHC recognition appears minimal.
- Department of Microbiology and Immunology, University of Melbourne, Parkville 3010, Victoria, Australia.
Organizational Affiliation: