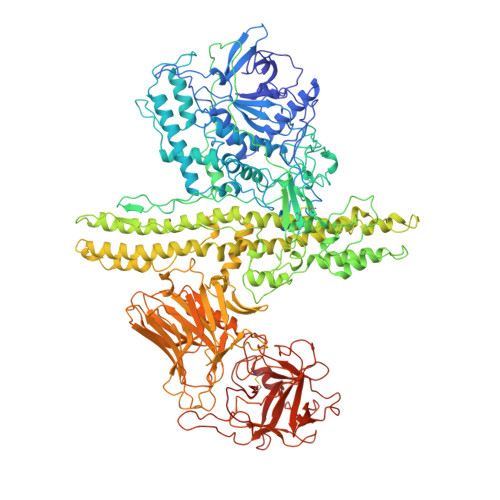
Crystal structure of botulinum neurotoxin type A and implications for toxicity.
Lacy, D.B., Tepp, W., Cohen, A.C., DasGupta, B.R., Stevens, R.C.(1998) Nat Struct Biol 5: 898-902
- PubMed: 9783750
- DOI: https://doi.org/10.1038/2338
- Primary Citation of Related Structures:
3BTA - PubMed Abstract:
Botulinum neurotoxin type A (BoNT/A) is the potent disease agent in botulism, a potential biological weapon and an effective therapeutic drug for involuntary muscle disorders. The crystal structure of the entire 1,285 amino acid di-chain neurotoxin was determined at 3.3 A resolution. The structure reveals that the translocation domain contains a central pair of alpha-helices 105 A long and a approximately 50 residue loop or belt that wraps around the catalytic domain. This belt partially occludes a large channel leading to a buried, negative active site--a feature that calls for radically different inhibitor design strategies from those currently used. The fold of the translocation domain suggests a mechanism of pore formation different from other toxins. Lastly, the toxin appears as a hybrid of varied structural motifs and suggests a modular assembly of functional subunits to yield pathogenesis.
- Department of Chemistry and Earnest Orlando Lawrence Berkeley National Laboratory, University of California, Berkeley, 94720, USA.
Organizational Affiliation: