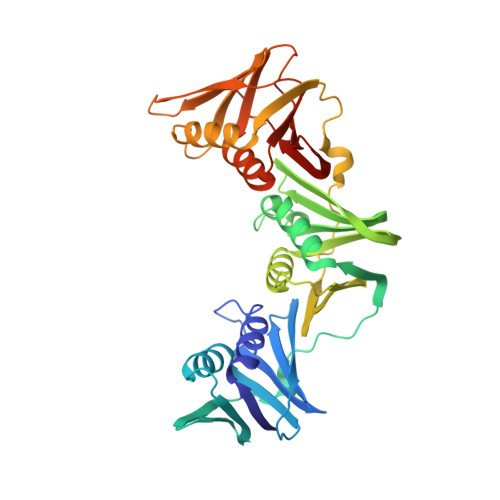
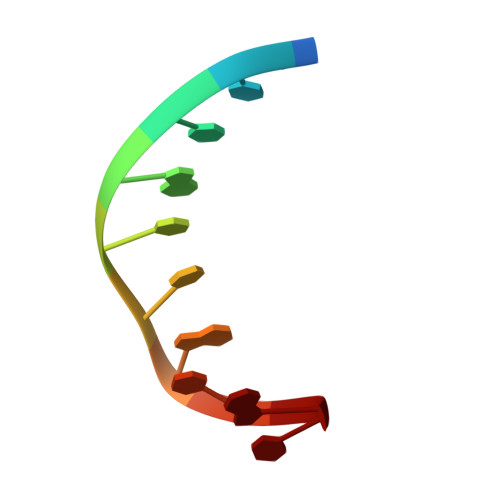
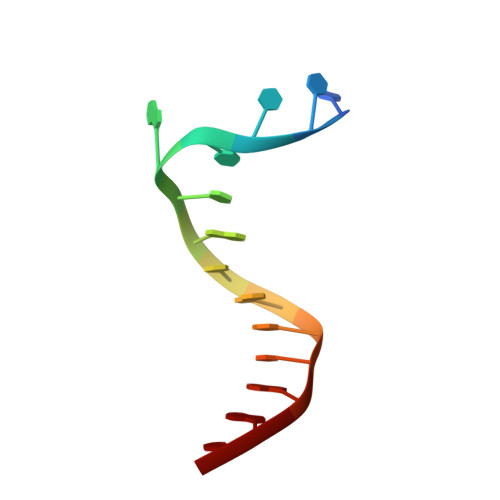
Structure of a sliding clamp on DNA
Georgescu, R.E., Kim, S.S., Yurieva, O., Kuriyan, J., Kong, X.-P., O'Donnell, M.(2008) Cell 132: 43-54
- PubMed: 18191219
- DOI: https://doi.org/10.1016/j.cell.2007.11.045
- Primary Citation of Related Structures:
3BEP - PubMed Abstract:
The structure of the E. coli beta clamp polymerase processivity factor has been solved in complex with primed DNA. Interestingly, the clamp directly binds the DNA duplex and also forms a crystal contact with the ssDNA template strand, which binds into the protein-binding pocket of the clamp. We demonstrate that these clamp-DNA interactions function in clamp loading, perhaps by inducing the ring to close around DNA. Clamp binding to template ssDNA may also serve to hold the clamp at a primed site after loading or during switching of multiple factors on the clamp. Remarkably, the DNA is highly tilted as it passes through the beta ring. The pronounced 22 degrees angle of DNA through beta may enable DNA to switch between multiple factors bound to a single clamp simply by alternating from one protomer of the ring to the other.
- Howard Hughes Medical Institute, Rockefeller University, 1230 York Avenue, Box 228, New York, NY 10021, USA.
Organizational Affiliation: