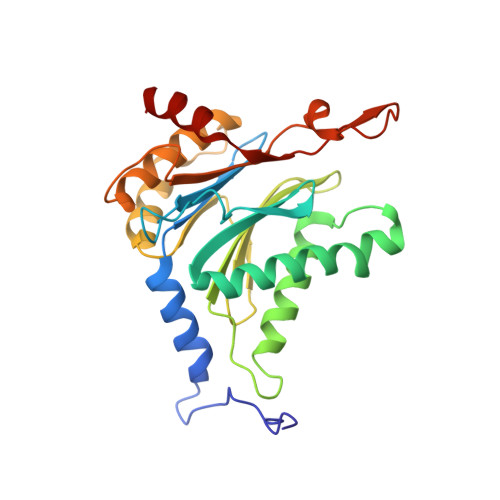
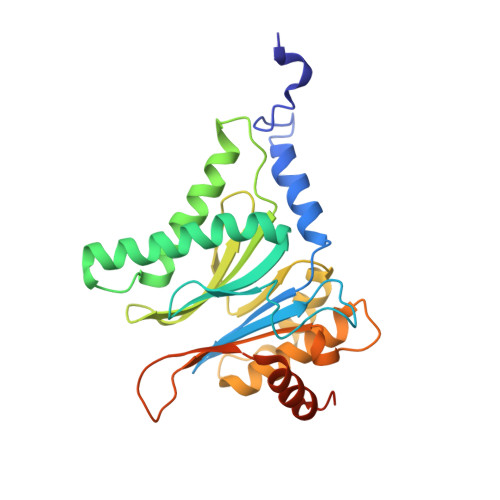
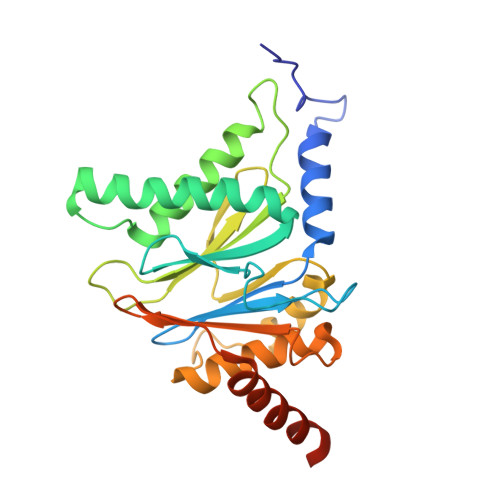
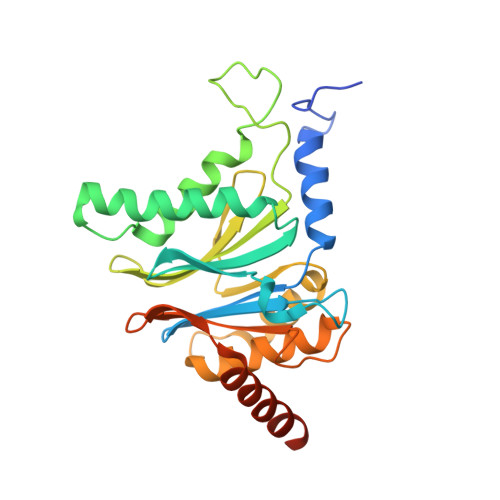
A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Groll, M., Schellenberg, B., Bachmann, A.S., Archer, C.R., Huber, R., Powell, T.K., Lindow, S., Kaiser, M., Dudler, R.(2008) Nature 452: 755-758
- PubMed: 18401409
- DOI: https://doi.org/10.1038/nature06782
- Primary Citation of Related Structures:
2ZCY, 3BDM - PubMed Abstract:
Pathogenic bacteria often use effector molecules to increase virulence. In most cases, the mode of action of effectors remains unknown. Strains of Pseudomonas syringae pv. syringae (Pss) secrete syringolin A (SylA), a product of a mixed non-ribosomal peptide/polyketide synthetase, in planta. Here we identify SylA as a virulence factor because a SylA-negative mutant in Pss strain B728a obtained by gene disruption was markedly less virulent on its host, Phaseolus vulgaris (bean). We show that SylA irreversibly inhibits all three catalytic activities of eukaryotic proteasomes, thus adding proteasome inhibition to the repertoire of modes of action of virulence factors. The crystal structure of the yeast proteasome in complex with SylA revealed a novel mechanism of covalent binding to the catalytic subunits. Thus, SylA defines a new class of proteasome inhibitors that includes glidobactin A (GlbA), a structurally related compound from an unknown species of the order Burkholderiales, for which we demonstrate a similar proteasome inhibition mechanism. As proteasome inhibitors are a promising class of anti-tumour agents, the discovery of a novel family of inhibitory natural products, which we refer to as syrbactins, may also have implications for the development of anti-cancer drugs. Homologues of SylA and GlbA synthetase genes are found in some other pathogenic bacteria, including the human pathogen Burkholderia pseudomallei, the causative agent of melioidosis. It is thus possible that these bacteria are capable of producing proteasome inhibitors of the syrbactin class.
- Center for Integrated Protein Science at the Department Chemie, Lehrstuhl für Biochemie, Technische Universität München, Lichtenbergstrasse 4, Garching D-85747, Germany.
Organizational Affiliation: