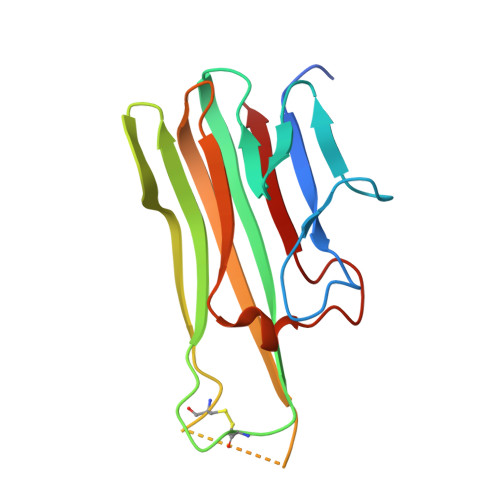
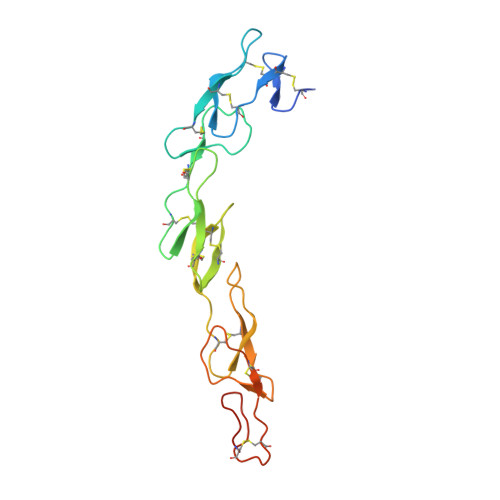
Solution of the Structure of the TNF-TNFR2 Complex
Mukai, Y., Nakamura, T., Yoshikawa, M., Yoshioka, Y., Tsunoda, S.I., Nakagawa, S., Yamagata, Y., Tsutsumi, Y.(2010) Sci Signal 3: ra83-ra83
- PubMed: 21081755
- DOI: https://doi.org/10.1126/scisignal.2000954
- Primary Citation of Related Structures:
3ALQ - PubMed Abstract:
Tumor necrosis factor (TNF) is an inflammatory cytokine that has important roles in various immune responses, which are mediated through its two receptors, TNF receptor 1 (TNFR1) and TNFR2. Antibody-based therapy against TNF is used clinically to treat several chronic autoimmune diseases; however, such treatment sometimes results in serious side effects, which are thought to be caused by the blocking of signals from both TNFRs. Therefore, knowledge of the structural basis for the recognition of TNF by each receptor would be invaluable in designing TNFR-selective drugs. Here, we solved the 3.0 angstrom resolution structure of the TNF-TNFR2 complex, which provided insight into the molecular recognition of TNF by TNFR2. Comparison to the known TNFR1 structure highlighted several differences between the ligand-binding interfaces of the two receptors. Additionally, we also demonstrated that TNF-TNFR2 formed aggregates on the surface of cells, which may be required for signal initiation. These results may contribute to the design of therapeutics for autoimmune diseases.
- Laboratory of Biopharmaceutical Research, National Institute of Biomedical Innovation, Osaka, Japan. yohe@phs.osaka-u.ac.jp
Organizational Affiliation: