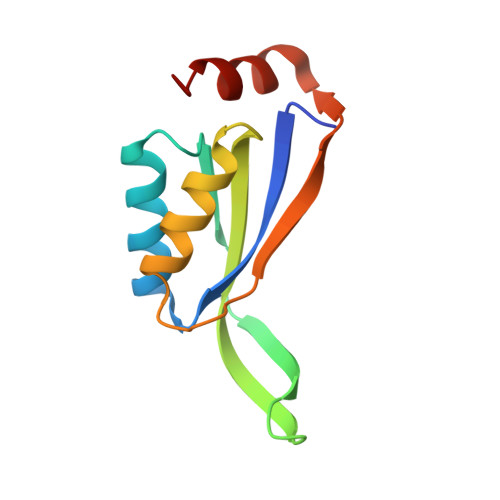
Crystal structure of stable protein CutA1 from psychrotrophic bacterium Shewanella sp. SIB1
Sato, A., Yokotani, S., Tadokoro, T., Tanaka, S.I., Angkawidjaja, C., Koga, Y., Takano, K., Kanaya, S.(2011) J Synchrotron Radiat 18: 6-10
- PubMed: 21169681
- DOI: https://doi.org/10.1107/S0909049510028669
- Primary Citation of Related Structures:
3AHP - PubMed Abstract:
CutA1 is widely found in bacteria, plants and animals, including humans. The functions of CutA1, however, have not been well clarified. It is known that CutA1s from Pyrococcus horikoshii, Thermus thermophilus and Oryza sativa unfold at temperatures remarkably higher than the growth temperatures of the host organisms. In this work the crystal structure of CutA1 from the psychrotrophic bacterium Shewanella sp. SIB1 (SIB1-CutA1) in a trimeric form was determined at 2.7 Å resolution. This is the first crystal structure of a psychrotrophic CutA1. The overall structure of SIB1-CutA1 is similar to those of CutA1 from Homo sapiens, Escherichia coli, Pyrococcus horikoshii, Thermus thermophilus, Termotoga maritima, Oryza sativa and Rattus norvergicus. A peculiarity is observed in the β2 strand. The β2 strand is divided into two short β strands, β2a and β2b, in SIB1-CutA1. A thermal denaturation experiment revealed that SIB1-CutA1 does not unfold completely at 363 K at pH 7.0, although Shewanella sp. SIB1 cannot grow at temperatures exceeding 303 K. These results indicate that the trimeric structural motif of CutA1 is the critical factor in its unusually high stability and suggest that CutA1 needs to maintain its high stability in order to function, even in psychrotrophs.
- Department of Material and Life Science, Osaka University, 2-1 Yamadaoka, Suita 565-0871, Japan.
Organizational Affiliation: