Two distinct mechanisms for the regulation of actin capping protein-competitive or allosteric inhibitions
Takeda, S., Minakata, S., Narita, A., Kitazawa, M., Yamakuni, T., Maeda, Y., Nitanai, Y.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| F-actin-capping protein subunit alpha-1 | 286 | Gallus gallus | Mutation(s): 0 Gene Names: CAPZA1 | 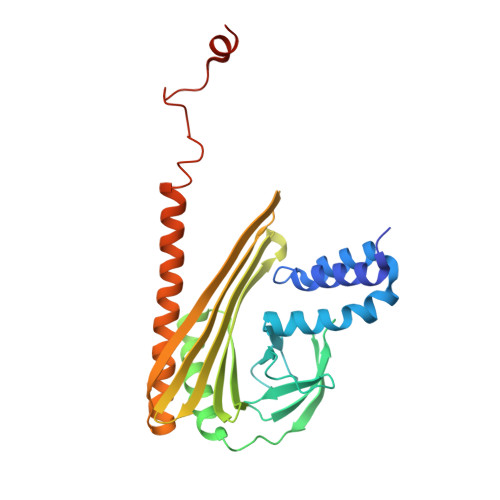 | |
UniProt | |||||
Find proteins for P13127 (Gallus gallus) Explore P13127 Go to UniProtKB: P13127 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13127 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| F-actin-capping protein subunit beta isoforms 1 and 2 | 277 | Gallus gallus | Mutation(s): 0 Gene Names: CAPZB | 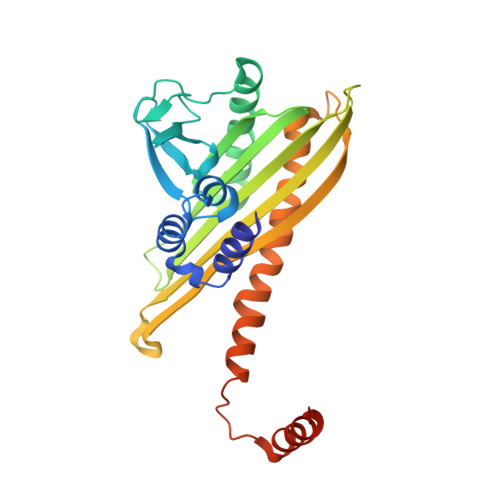 | |
UniProt | |||||
Find proteins for P14315 (Gallus gallus) Explore P14315 Go to UniProtKB: P14315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14315 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 32mer peptide from Leucine-rich repeat-containing protein 16A | K [auth X], L [auth Y], M [auth Z], N [auth W], O [auth V] | 37 | Mus musculus | Mutation(s): 0 Gene Names: Lrrc16a |  |
UniProt | |||||
Find proteins for Q6EDY6 (Mus musculus) Explore Q6EDY6 Go to UniProtKB: Q6EDY6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6EDY6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 231.153 | α = 90 |
| b = 104.358 | β = 119.09 |
| c = 186.633 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| DPS | data reduction |
| HKL-2000 | data scaling |