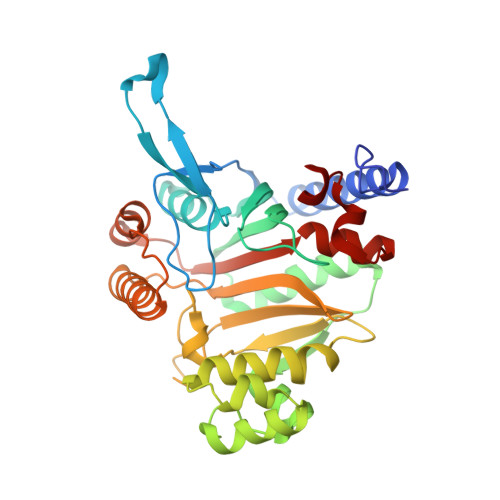
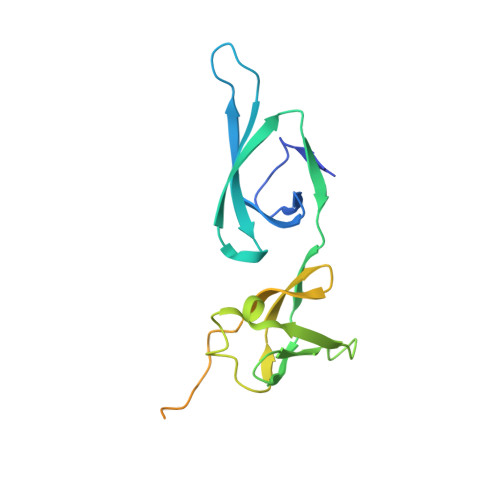
A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Yanagisawa, T., Sumida, T., Ishii, R., Takemoto, C., Yokoyama, S.(2010) Nat Struct Mol Biol 17: 1136-1143
- PubMed: 20729861
- DOI: https://doi.org/10.1038/nsmb.1889
- Primary Citation of Related Structures:
3A5Y, 3A5Z - PubMed Abstract:
Aminoacyl-tRNA synthetase (aaRS) paralogs with unknown functions exist in various species. We now report novel 'protein lysylation' by an Escherichia coli lysyl-tRNA synthetase paralog, GenX/PoxA/YjeA. X-ray crystallographic analysis shows that the structure of the GenX protein resembles that of a class II aaRS. Further in vitro studies reveal that it specifically aminoacylates EF-P with lysine. The shape of the protein substrate mimics that of the L-shaped tRNA, and its lysylation site corresponds to the tRNA 3' end. Thus, we show how the aaRS architecture can be adapted to achieve aminoacylation of a specific protein. Moreover, in vivo analyses reveal that the translation elongation factor P (EF-P) lysylation by GenX is enhanced by YjeK (lysine 2,3-aminomutase paralog), which is encoded next to the EF-P gene, and might convert alpha-lysyl-EF-P to beta-lysyl-EF-P. In vivo analyses indicate that the EF-P modification by GenX and YjeK is essential for cell survival.
- RIKEN Systems and Structural Biology Center, Tsurumi, Yokohama, Japan.
Organizational Affiliation: