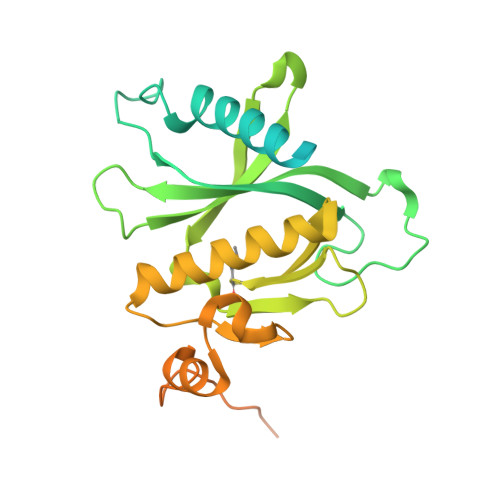
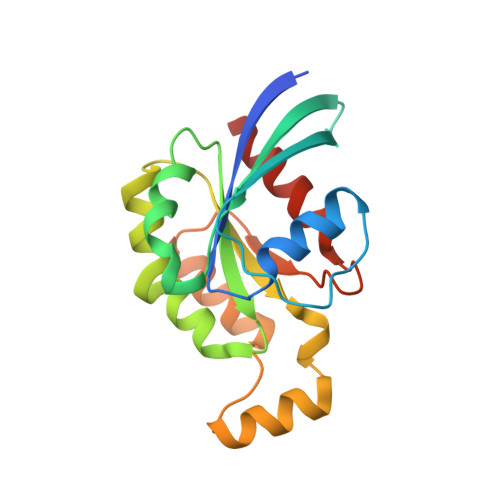
Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3
Yamashita, M., Kurokawa, K., Sato, Y., Yamagata, A., Mimura, H., Yoshikawa, A., Sato, K., Nakano, A., Fukai, S.(2010) Nat Struct Mol Biol 17: 180-186
- PubMed: 20062059
- DOI: https://doi.org/10.1038/nsmb.1722
- Primary Citation of Related Structures:
3A58 - PubMed Abstract:
The exocyst complex is a hetero-octameric protein complex that functions during cell polarization by tethering the secretory vesicle to the target membrane. The yeast exocyst subunit Sec3 binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P(2)) and the small GTPases Rho1 and Cdc42 via its N-terminal domain (Sec3-N), and these interactions target Sec3 to the plasma membrane. Here we report the crystal structure of the Sec3-N in complex with Rho1 at 2.6-A resolution. Sec3-N adopts a pleckstrin homology (PH) fold, despite having no detectable sequence homology with other PH domains of known structure. Clusters of conserved basic residues constitute a positively charged cleft, which was identified as a binding site for PtdIns(4,5)P(2). Residues Phe77, Ile115 and Leu131 of Sec3 bind to an extended hydrophobic surface formed around switch regions I and II of Rho1. To our knowledge, these are the first structural insights into how an exocyst subunit might interact with both protein and phospholipid factors on the target membrane.
- Structural Biology Laboratory, Life Science Division, Synchrotron Radiation Research Organization and Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: