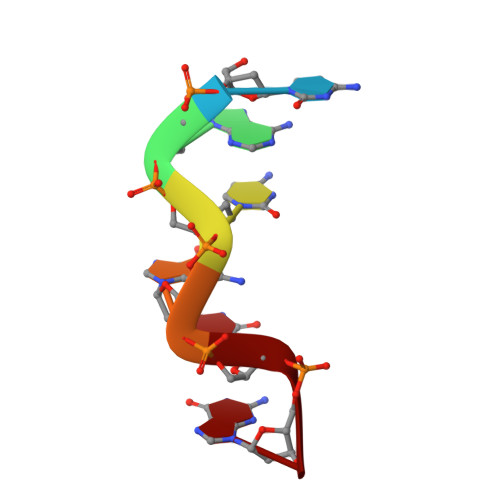
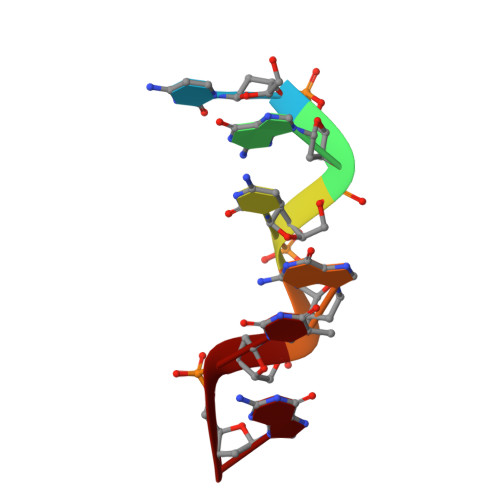
Structure of d(CACGCG).d(CGCGTG) in crystals grown in the presence of ruthenium III hexammine chloride.
Karthe, P., Gautham, N.(1998) Acta Crystallogr D Biol Crystallogr 54: 501-509
- PubMed: 9761846
- DOI: https://doi.org/10.1107/s0907444997013954
- Primary Citation of Related Structures:
351D - PubMed Abstract:
Hexammine ions are strong inducers of the transition from the B-form to the left-handed Z-form in DNA. Here the structure of d(CACGCG). d(CGCGTG) obtained from crystals grown from a drop containing [Ru(NH3)6]Cl3 is reported. The structure is clearly characterized as Z-DNA. When compared with the structure of d(CACGCG).d(CGCGTG)/MgCl2 and that of d(CGCGCG)2, subtle differences are seen, most noticeably in the water structure. Since stable well diffracting crystals grow easily in the presence of [Ru(NH3)6]Cl3 and since this ion is not visible in the electron density it is concluded that the ion plays a non-specific role in stabilizing Z-DNA.
- Department of Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai 600 025, India. crystal@giasmd01.vsnl.net.in
Organizational Affiliation: