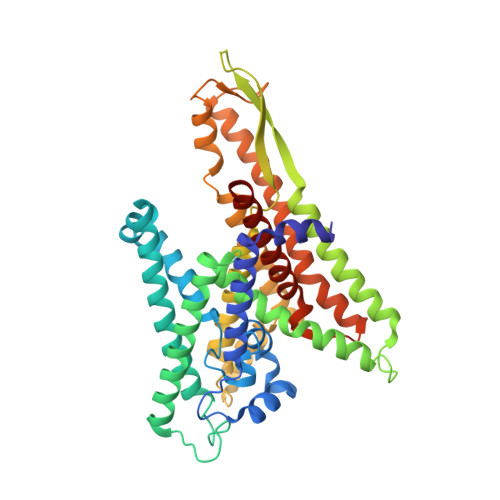
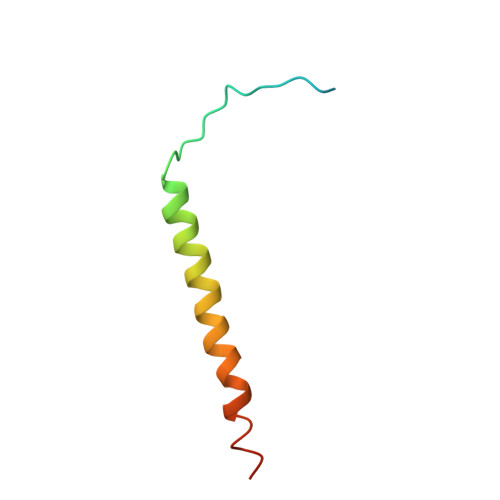
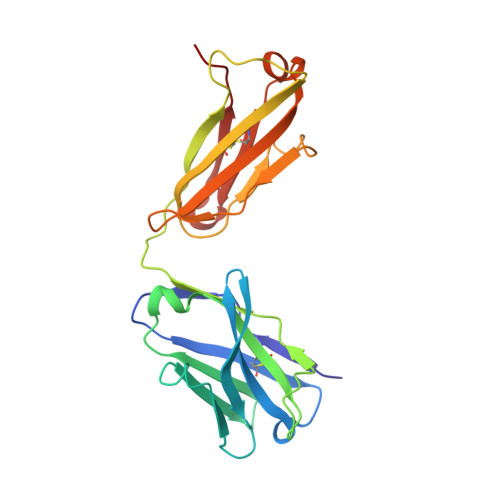
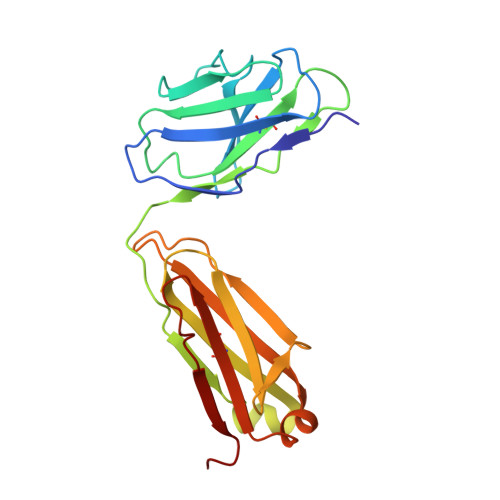
Conformational transition of Sec machinery inferred from bacterial SecYE structures
Tsukazaki, T., Mori, H., Fukai, S., Ishitani, R., Mori, T., Dohmae, N., Perederina, A., Sugita, Y., Vassylyev, D.G., Ito, K., Nureki, O.(2008) Nature 455: 988-991
- PubMed: 18923527
- DOI: https://doi.org/10.1038/nature07421
- Primary Citation of Related Structures:
2ZJS, 2ZQP - PubMed Abstract:
Over 30% of proteins are secreted across or integrated into membranes. Their newly synthesized forms contain either cleavable signal sequences or non-cleavable membrane anchor sequences, which direct them to the evolutionarily conserved Sec translocon (SecYEG in prokaryotes and Sec61, comprising alpha-, gamma- and beta-subunits, in eukaryotes). The translocon then functions as a protein-conducting channel. These processes of protein localization occur either at or after translation. In bacteria, the SecA ATPase drives post-translational translocation. The only high-resolution structure of a translocon available so far is that for SecYEbeta from the archaeon Methanococcus jannaschii, which lacks SecA. Here we present the 3.2-A-resolution crystal structure of the SecYE translocon from a SecA-containing organism, Thermus thermophilus. The structure, solved as a complex with an anti-SecY Fab fragment, revealed a 'pre-open' state of SecYE, in which several transmembrane helices are shifted, as compared to the previous SecYEbeta structure, to create a hydrophobic crack open to the cytoplasm. Fab and SecA bind to a common site at the tip of the cytoplasmic domain of SecY. Molecular dynamics and disulphide mapping analyses suggest that the pre-open state might represent a SecYE conformational transition that is inducible by SecA binding. Moreover, we identified a SecA-SecYE interface that comprises SecA residues originally buried inside the protein, indicating that both the channel and the motor components of the Sec machinery undergo cooperative conformational changes on formation of the functional complex.
- Department of Biological Information, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama-shi, Kanagawa 226-8501, Japan.
Organizational Affiliation: