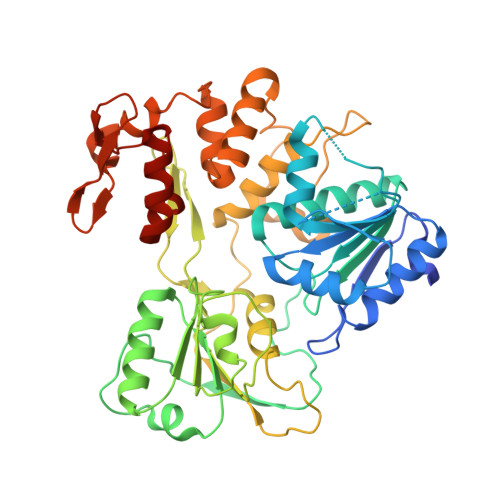
Crystal Structure of Catalytic Domain of Japanese Encephalitis Virus NS3 Helicase/Nucleoside Triphosphatase at a Resolution 1.8
Yamashita, T., Unno, H., Mori, Y., Tani, H., Moriishi, K., Takamizawa, A., Agoh, M., Tsukihara, T., Matsuura, Y.To be published.