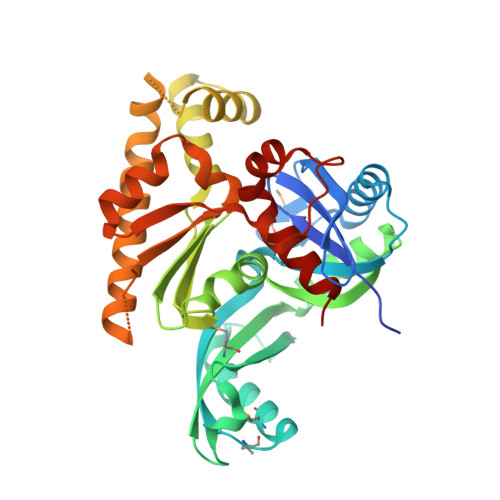
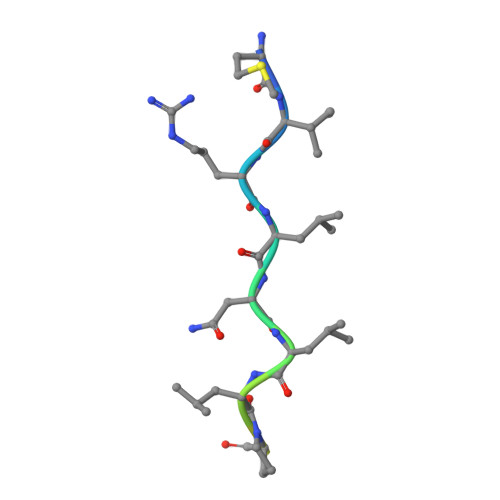
Structure of the Pilm-Piln Inner Membrane Type Iv Pilus Biogenesis Complex from Thermus Thermophilus.
Karuppiah, V., Derrick, J.P.(2011) J Biological Chem 286: 24434
- PubMed: 21596754
- DOI: https://doi.org/10.1074/jbc.M111.243535
- Primary Citation of Related Structures:
2YCH - PubMed Abstract:
Type IV pili are surface-exposed filaments, which extend from a variety of bacterial pathogens and play a major role in pathogenesis, motility, and DNA uptake. Here, we present the crystal structure of a complex between a cytoplasmic component of the type IV pilus biogenesis system from Thermus thermophilus, PilM, in complex with a peptide derived from the cytoplasmic portion of the inner membrane protein PilN. PilM also binds ATP, and its structure is most similar to the actin-like protein FtsA. PilN binds in a narrow channel between the 1A and 1C subdomains in PilM; the binding site is well conserved in other gram-negative bacteria, notably Neisseria meningitidis, Pseudomonas aeruginosa, and Vibrio cholerae. We find no evidence for the catalysis of ATP hydrolysis by PilM; fluorescence data indicate that the protein is likely to be saturated by ATP at physiological concentrations. In addition, binding of the PilN peptide appears to influence the environment of the ATP binding site. This is the first reported structure of a complex between two type IV pilus biogenesis proteins. We propose a model in which PilM binds ATP and then PilN as one of the first steps in the formation of the inner membrane platform of the type IV pilus biogenesis complex.
- Faculty of Life Sciences, The University of Manchester, Oxford Road, Manchester M13 9PT, United Kingdom.
Organizational Affiliation: