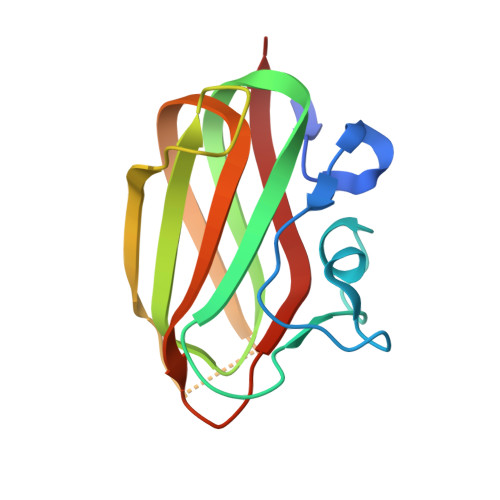
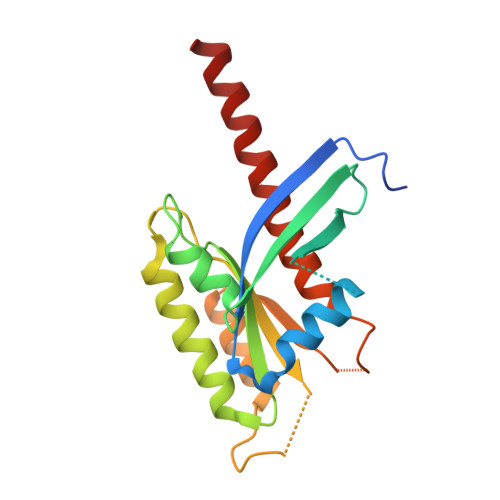
Crystal Structure of the Intraflagellar Transport Complex 25/27.
Bhogaraju, S., Taschner, M., Morawetz, M., Basquin, C., Lorentzen, E.(2011) EMBO J 30: 1907
- PubMed: 21505417
- DOI: https://doi.org/10.1038/emboj.2011.110
- Primary Citation of Related Structures:
2YC2, 2YC4 - PubMed Abstract:
The cilium is an important organelle that is found on many eukaryotic cells, where it serves essential functions in motility, sensory reception and signalling. Intraflagellar transport (IFT) is a vital process for the formation and maintenance of cilia. We have determined the crystal structure of Chlamydomonas reinhardtii IFT25/27, an IFT sub-complex, at 2.6 Å resolution. IFT25 and IFT27 interact via a conserved interface that we verify biochemically using structure-guided mutagenesis. IFT27 displays the fold of Rab-like small guanosine triphosphate hydrolases (GTPases), binds GTP and GDP with micromolar affinity and has very low intrinsic GTPase activity, suggesting that it likely requires a GTPase-activating protein (GAP) for robust GTP turnover. A patch of conserved surface residues contributed by both IFT25 and IFT27 is found adjacent to the GTP-binding site and could mediate the binding to other IFT proteins as well as to a potential GAP. These results provide the first step towards a high-resolution structural understanding of the IFT complex.
- Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: