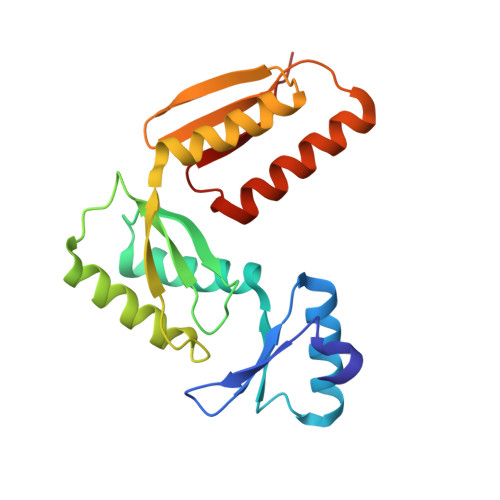
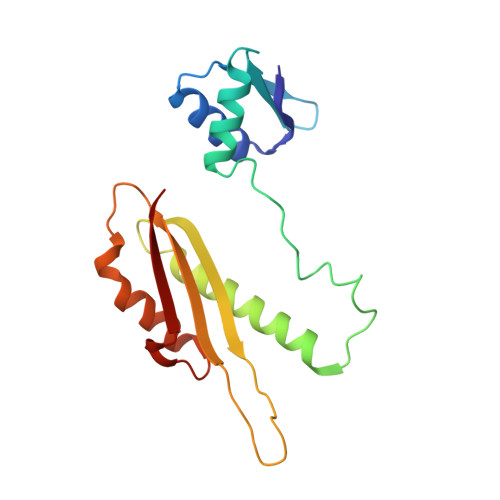
Three-Dimensional Model of Salmonella'S Needle Complex at Subnanometer Resolution.
Schraidt, O., Marlovits, T.C.(2011) Science 331: 1192-1195
- PubMed: 21385715
- DOI: https://doi.org/10.1126/science.1199358
- Primary Citation of Related Structures:
2Y9J, 2Y9K - PubMed Abstract:
Type III secretion systems (T3SSs) are essential virulence factors used by many Gram-negative bacteria to inject proteins that make eukaryotic host cells accessible to invasion. The T3SS core structure, the needle complex (NC), is a ~3.5 megadalton-sized, oligomeric, membrane-embedded complex. Analyzing cryo-electron microscopy images of top views of NCs or NC substructures from Salmonella typhimurium revealed a 24-fold symmetry for the inner rings and a 15-fold symmetry for the outer rings, giving an overall C3 symmetry. Local refinement and averaging showed the organization of the central core and allowed us to reconstruct a subnanometer composite structure of the NC, which together with confident docking of atomic structures reveal insights into its overall organization and structural requirements during assembly.
- Research Institute of Molecular Pathology, Dr. Bohr Gasse 7, A-1030 Vienna, Austria.
Organizational Affiliation: