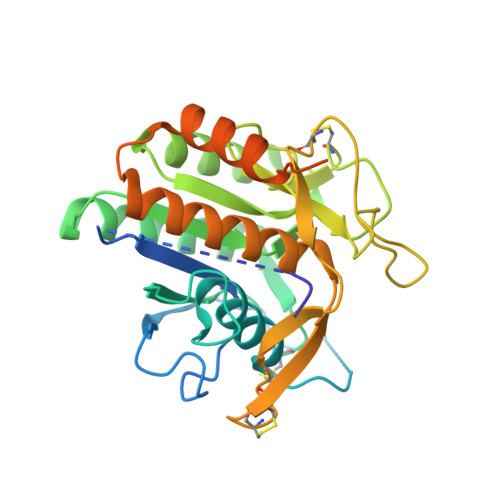
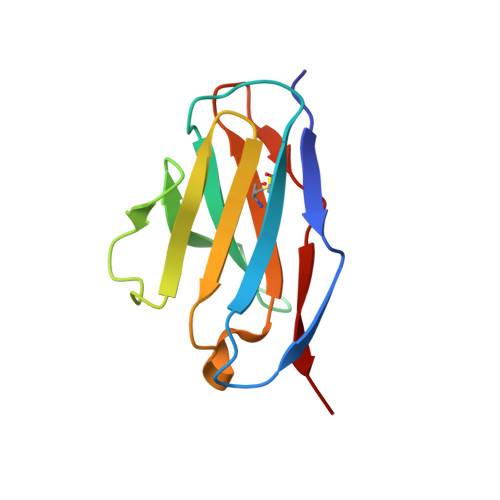
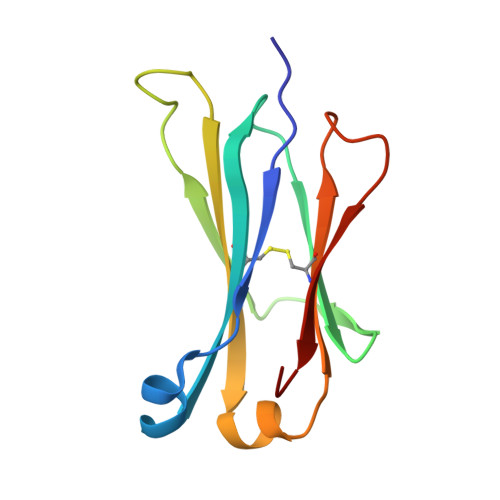
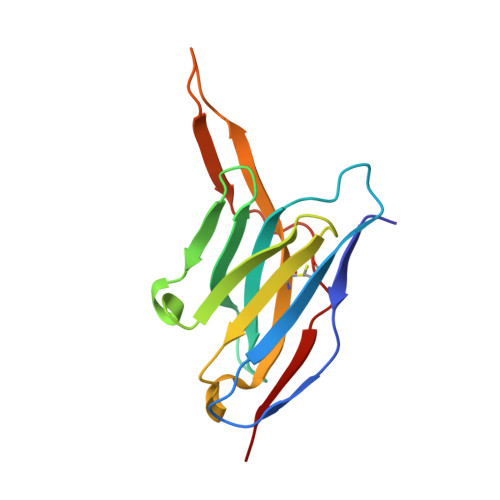
A Pcsk9-Binding Antibody that Structurally Mimics the Egf(A) Domain of Ldl-Receptor Reduces Ldl Cholesterol in Vivo.
Ni, Y.G., Di Marco, S., Condra, J.H., Peterson, L.B., Wang, W., Wang, F., Pandit, S., Hammond, H.A., Rosa, R., Cummings, R.T., Wood, D.D., Liu, X., Bottomley, M.J., Shen, X., Cubbon, R.M., Wang, S.P., Johns, D.G., Volpari, C., Hamuro, L., Chin, J., Huang, L., Zhao, J.Z., Vitelli, S., Haytko, P., Wisniewski, D., Mitnaul, L.J., Sparrow, C.P., Hubbard, B., Carfi, A., Sitlani, A.(2011) J Lipid Res 52: 78
- PubMed: 20959675
- DOI: https://doi.org/10.1194/jlr.M011445
- Primary Citation of Related Structures:
2XTJ - PubMed Abstract:
Proprotein convertase subtilisin-like/kexin type 9 (PCSK9) regulates LDL cholesterol levels by inhibiting LDL receptor (LDLr)-mediated cellular LDL uptake. We have identified a fragment antigen-binding (Fab) 1D05 which binds PCSK9 with nanomolar affinity. The fully human antibody 1D05-IgG2 completely blocks the inhibitory effects of wild-type PCSK9 and two gain-of-function human PCSK9 mutants, S127R and D374Y. The crystal structure of 1D05-Fab bound to PCSK9 reveals that 1D05-Fab binds to an epitope on the PCSK9 catalytic domain which includes the entire LDLr EGF(A) binding site. Notably, the 1D05-Fab CDR-H3 and CDR-H2 loops structurally mimic the EGF(A) domain of LDLr. In a transgenic mouse model (CETP/LDLr-hemi), in which plasma lipid and PCSK9 profiles are comparable to those of humans, 1D05-IgG2 reduces plasma LDL cholesterol to 40% and raises hepatic LDLr protein levels approximately fivefold. Similarly, in healthy rhesus monkeys, 1D05-IgG2 effectively reduced LDL cholesterol 20%-50% for over 2 weeks, despite its relatively short terminal half-life (t(1/2) = 3.2 days). Importantly, the decrease in circulating LDL cholesterol corresponds closely to the reduction in free PCSK9 levels. Together these results clearly demonstrate that the LDL-lowering effect of the neutralizing anti-PCSK9 1D05-IgG2 antibody is mediated by reducing the amount of PCSK9 that can bind to the LDLr.
- Department of Cardiovascular Diseases, Merck Research Laboratories, Rahway, New Jersey, USA. yan_ni@merck.com
Organizational Affiliation: