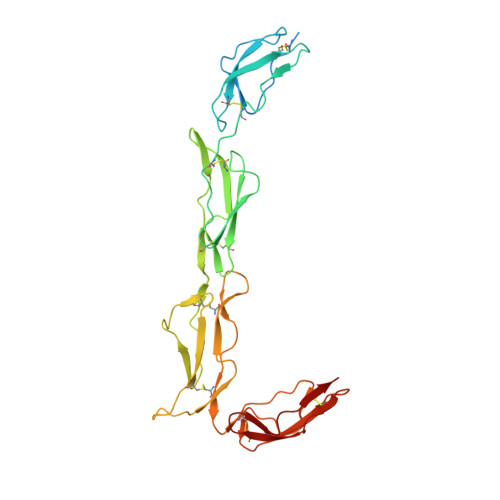
Structures of the Rat Complement Regulator Crry.
Roversi, P., Johnson, S., Caesar, J.J.E., Mclean, F., Leath, K.J., Tsiftsoglou, S.A., Morgan, B.P., Harris, C.L., Sim, R.B., Lea, S.M.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 739
- PubMed: 21795784
- DOI: https://doi.org/10.1107/S1744309111016551
- Primary Citation of Related Structures:
2XRB, 2XRD - PubMed Abstract:
Complement receptor 1-related protein Y (CrrY) is an important cell-surface regulator of complement that is unique to rodent species. The structure of rat CrrY domains 1-4 has been determined in two distinct crystal forms and reveals a 70° bend between domains 3 and 4. Comparisons of this structure with those of other complement regulators suggests that rearrangement of this interface may occur on forming the regulatory complex with C3b.
- Sir William Dunn School of Pathology, Oxford University, South Parks Road, Oxford OX4 3RE, England.
Organizational Affiliation: