The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Leo, J.C., Lyskowski, A., Hattula, K., Hartmann, M.D., Schwarz, H., Butcher, S.J., Linke, D., Lupas, A.N., Goldman, A.(2011) Structure 19: 1021
- PubMed: 21742268
- DOI: https://doi.org/10.1016/j.str.2011.03.021
- Primary Citation of Related Structures:
2XQH, 2XZR - PubMed Abstract:
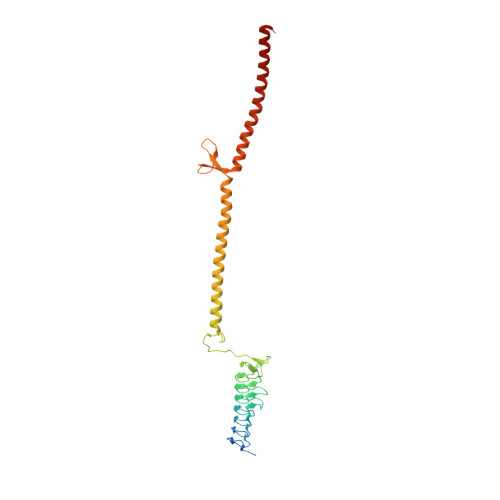
The Escherichia coli Ig-binding (Eib) proteins are trimeric autotransporter adhesins (TAAs) and receptors for IgG Fc. We present the structure of a large fragment of the passenger domain of EibD, the first TAA structure to have both a YadA-like head domain and the entire coiled-coil stalk. The stalk begins as a right-handed superhelix, but switches handedness halfway down. An unexpected β-minidomain joins the two and inserts a ∼120° rotation such that there is no net twist between the beginning and end of the stalk. This may be important in folding and autotransport. The surprisingly large cavities we found in EibD and other TAAs may explain how TAAs bend to bind their ligands. We identified how IgA and IgG bind and modeled the EibD-IgG Fc complex. We further show that EibD promotes autoagglutination and biofilm formation and forms a fibrillar layer covering the cell surface making zipper-like contacts between cells.
- Macromolecular Crystallography Group, Institute of Biotechnology, University of Helsinki, 00014 Helsinki, Finland.
Organizational Affiliation: