Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Wohlkonig, A., Chan, P.F., Fosberry, A.P., Homes, P., Huang, J., Kranz, M., Leydon, V.R., Miles, T.J., Pearson, N.D., Perera, R.L., Shillings, A.J., Gwynn, M.N., Bax, B.D.(2010) Nat Struct Mol Biol 17: 1152
- PubMed: 20802486
- DOI: https://doi.org/10.1038/nsmb.1892
- Primary Citation of Related Structures:
2XKJ, 2XKK - PubMed Abstract:
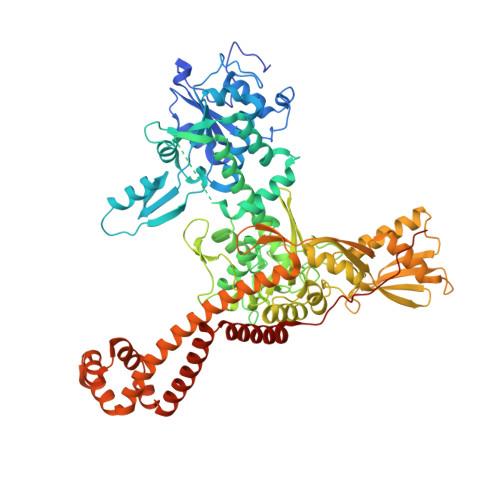
Quinolone antibacterials have been used to treat bacterial infections for over 40 years. A crystal structure of moxifloxacin in complex with Acinetobacter baumannii topoisomerase IV now shows the wedge-shaped quinolone stacking between base pairs at the DNA cleavage site and binding conserved residues in the DNA cleavage domain through chelation of a noncatalytic magnesium ion. This provides a molecular basis for the quinolone inhibition mechanism, resistance mutations and invariant quinolone antibacterial structural features.
- Platform Technology and Science, GlaxoSmithKline, Medicines Research Centre, Stevenage, Hertfordshire, UK.
Organizational Affiliation: