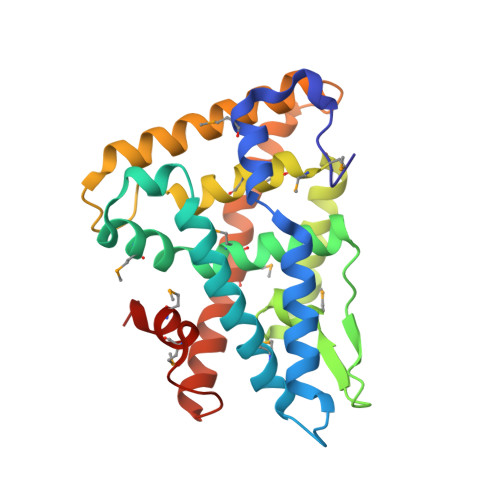
Crystal Structure of Fushi Tarazu Factor 1 Ligand Binding Domain/Fushi Tarazu Peptide Complex Identifies New Class of Nuclear Receptors.
Yoo, J., Ko, S., Kim, H., Sampson, H., Yun, J.H., Choe, K.M., Chang, I., Arrowsmith, C.H., Krause, H.M., Cho, H.S., Lee, W.(2011) J Biological Chem 286: 31225
- PubMed: 21775434
- DOI: https://doi.org/10.1074/jbc.M111.252916
- Primary Citation of Related Structures:
2XHS - PubMed Abstract:
The interaction between the orphan nuclear receptor FTZ-F1 (Fushi tarazu factor 1) and the segmentation gene protein FTZ is critical for specifying alternate parasegments in the Drosophila embryo. Here, we have determined the structure of the FTZ-F1 ligand-binding domain (LBD)·FTZ peptide complex using x-ray crystallography. Strikingly, the ligand-binding pocket of the FTZ-F1 LBD is completely occupied by helix 6 (H6) of the receptor, whereas the cofactor FTZ binds the co-activator cleft site of the FTZ-F1 LBD. Our findings suggest that H6 is essential for transcriptional activity of FTZ-F1; this is further supported by data from mutagenesis and activity assays. These data suggest that FTZ-F1 might belong to a novel class of ligand-independent nuclear receptors. Our findings are intriguing given that the highly homologous human steroidogenic factor-1 and liver receptor homolog-1 LBDs exhibit sizable ligand-binding pockets occupied by putative ligand molecules.
- Department of Biology, College of Life Science and Biotechnology, Yonsei University, Shinchon-dong, Seodaemun-gu 134, Seoul 120-749, Korea.
Organizational Affiliation: