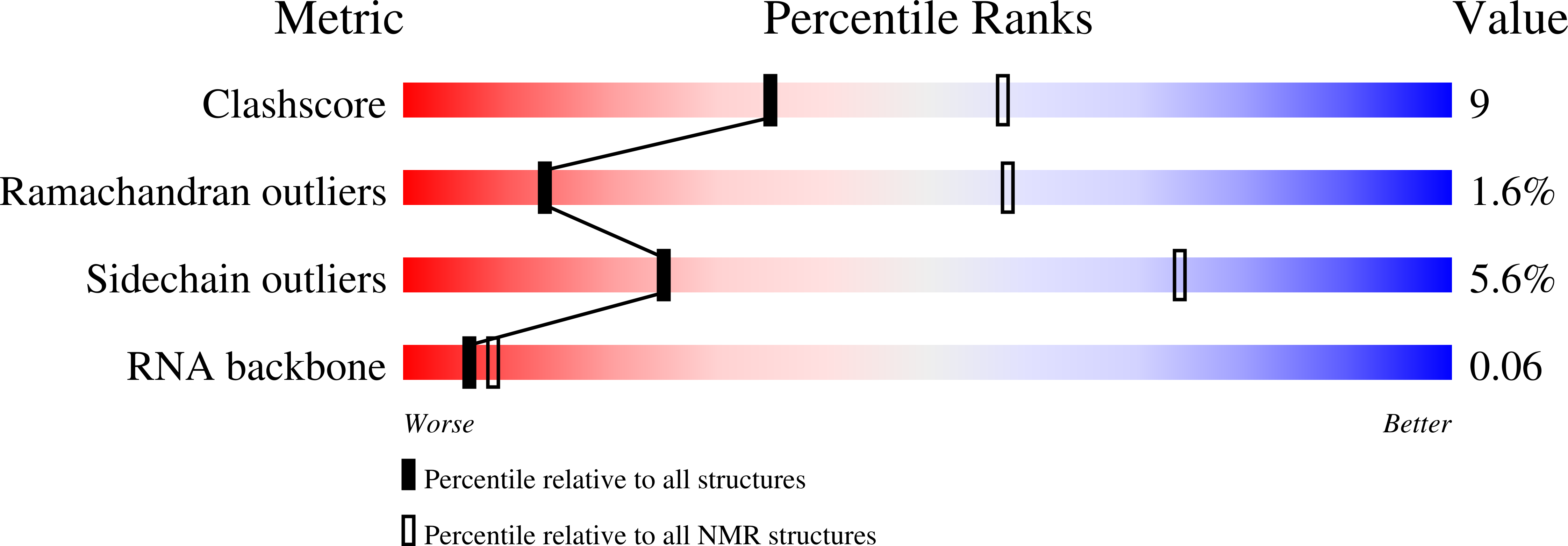
Recognition of 2'-O-Methylated 3'-End of Pirna by the Paz Domain of a Piwi Protein.
Simon, B., Kirkpatrick, J.P., Eckhardt, S., Reuter, M., Rocha, E.A., Andrade-Navarro, M.A., Sehr, P., Pillai, R.S., Carlomagno, T.(2011) Structure 19: 172
- PubMed: 21237665
- DOI: https://doi.org/10.1016/j.str.2010.11.015
- Primary Citation of Related Structures:
2XFM - PubMed Abstract:
Piwi proteins are germline-specific Argonautes that associate with small RNAs called Piwi-interacting RNAs (piRNAs), and together with these RNAs are implicated in transposon silencing. The PAZ domain of Argonaute proteins recognizes the 3'-end of the RNA, which in the case of piRNAs is invariably modified with a 2'-O-methyl group. Here, we present the solution structure of the PAZ domain from the mouse Piwi protein, MIWI, in complex with an 8-mer piRNA mimic. The methyl group is positioned in a hydrophobic cavity made of conserved amino acids from strand β7 and helix α3, where it is contacted by the side chain of methionine-382. Our structure is similar to that of Ago-PAZ, but subtle differences illustrate how the PAZ domain has evolved to accommodate distinct 3' ends from a variety of RNA substrates.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.
Organizational Affiliation: