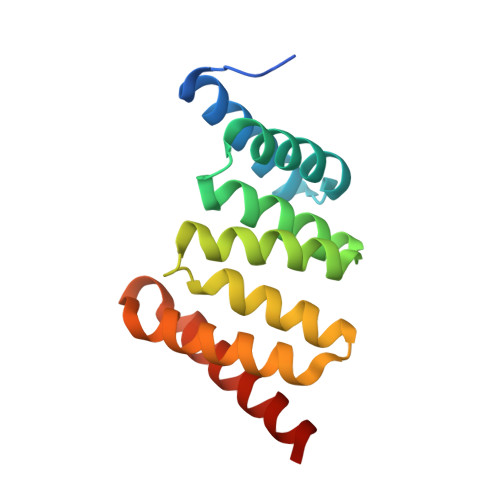
Structural Basis of Chaperone Recognition of Type III Secretion System Minor Translocator Proteins.
Job, V., Mattei, P.-J., Lemaire, D., Attree, I., Dessen, A.(2010) J Biological Chem 285: 23224
- PubMed: 20385547
- DOI: https://doi.org/10.1074/jbc.M110.111278
- Primary Citation of Related Structures:
2XCB, 2XCC - PubMed Abstract:
The type III secretion system (T3SS) is a complex nanomachine employed by many Gram-negative pathogens, including the nosocomial agent Pseudomonas aeruginosa, to inject toxins directly into the cytoplasm of eukaryotic cells. A key component of all T3SS is the translocon, a proteinaceous channel that is inserted into the target membrane, which allows passage of toxins into target cells. In most bacterial species, two distinct membrane proteins (the "translocators") are involved in translocon formation, whereas in the bacterial cytoplasm, however, they remain associated to a common chaperone. To date, the strategy employed by a single chaperone to recognize two distinct translocators is unknown. Here, we report the crystal structure of a complex between the Pseudomonas translocator chaperone PcrH and a short region from the minor translocator PopD. PcrH displays a 7-helical tetratricopeptide repeat fold that harbors the PopD peptide within its concave region, originally believed to be involved in recognition of the major translocator, PopB. Point mutations introduced into the PcrH-interacting region of PopD impede translocator-chaperone recognition in vitro and lead to impairment of bacterial cytotoxicity toward macrophages in vivo. These results indicate that T3SS translocator chaperones form binary complexes with their partner molecules, and the stability of their interaction regions must be strictly maintained to guarantee bacterial infectivity. The PcrH-PopD complex displays homologs among a number of pathogenic strains and could represent a novel, potential target for antibiotic development.
- Bacterial Pathogenesis Group, Institut de Biologie Structurale, UMR 5075, CNRS/Commissariat à l'Enérgie Atomique/Université Joseph Fourier, 41 Rue Jules Horowitz, 38027 Grenoble, France.
Organizational Affiliation: