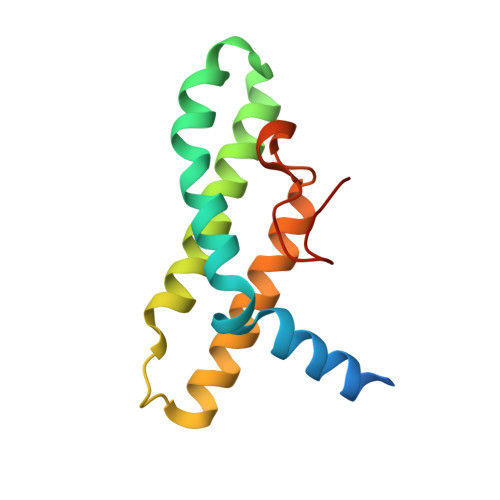
The Peroxisomal Receptor Pex19P Forms a Helical Mpts Recognition Domain.
Schueller, N., Holton, S.J., Fodor, K., Milewski, M., Konarev, P., Stanley, W.A., Wolf, J., Erdmann, R., Schliebs, W., Song, Y.H., Wilmanns, M.(2010) EMBO J 29: 2491
- PubMed: 20531392
- DOI: https://doi.org/10.1038/emboj.2010.115
- Primary Citation of Related Structures:
2WL8 - PubMed Abstract:
The protein Pex19p functions as a receptor and chaperone of peroxisomal membrane proteins (PMPs). The crystal structure of the folded C-terminal part of the receptor reveals a globular domain that displays a bundle of three long helices in an antiparallel arrangement. Complementary functional experiments, using a range of truncated Pex19p constructs, show that the structured alpha-helical domain binds PMP-targeting signal (mPTS) sequences with about 10 muM affinity. Removal of a conserved N-terminal helical segment from the mPTS recognition domain impairs the ability for mPTS binding, indicating that it forms part of the mPTS-binding site. Pex19p variants with mutations in the same sequence segment abolish correct cargo import. Our data indicate a divided N-terminal and C-terminal structural arrangement in Pex19p, which is reminiscent of a similar division in the Pex5p receptor, to allow separation of cargo-targeting signal recognition and additional functions.
- EMBL c/o DESY, Notkestrasse 85, Hamburg, Germany.
Organizational Affiliation: