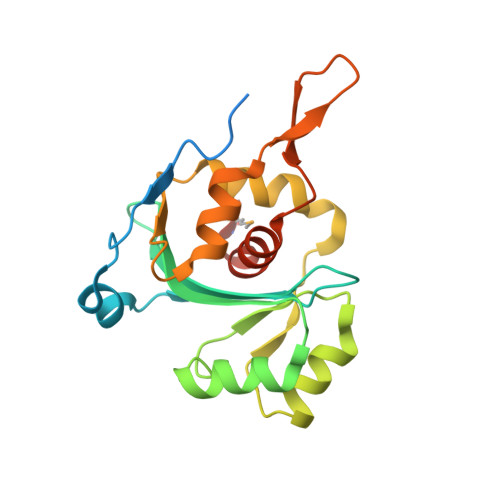
Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Smits, C., Chechik, M., Kovalevskiy, O.V., Shevtsov, M.B., Foster, A.W., Alonso, J.C., Antson, A.A.(2009) EMBO Rep 10: 592
- PubMed: 19444313
- DOI: https://doi.org/10.1038/embor.2009.53
- Primary Citation of Related Structures:
2WBN, 2WC9 - PubMed Abstract:
The DNA-packaging motor in tailed bacteriophages requires nuclease activity to ensure that the genome is packaged correctly. This nuclease activity is tightly regulated as the enzyme is inactive for the duration of DNA translocation. Here, we report the X-ray structure of the large terminase nuclease domain from bacteriophage SPP1. Similarity with the RNase H family endonucleases allowed interactions with the DNA to be predicted. A structure-based alignment with the distantly related T4 gp17 terminase shows the conservation of an extended beta-sheet and an auxiliary beta-hairpin that are not found in other RNase H family proteins. The model with DNA suggests that the beta-hairpin partly blocks the active site, and in vivo activity assays show that the nuclease domain is not functional in the absence of the ATPase domain. Here, we propose that the nuclease activity is regulated by movement of the beta-hairpin, altering active site access and the orientation of catalytically essential residues.
- York Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5YW, UK.
Organizational Affiliation: