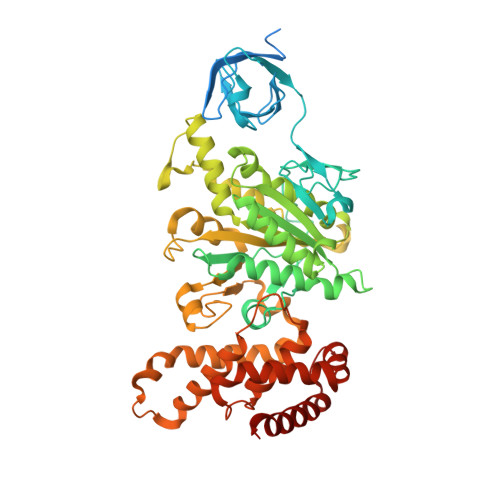
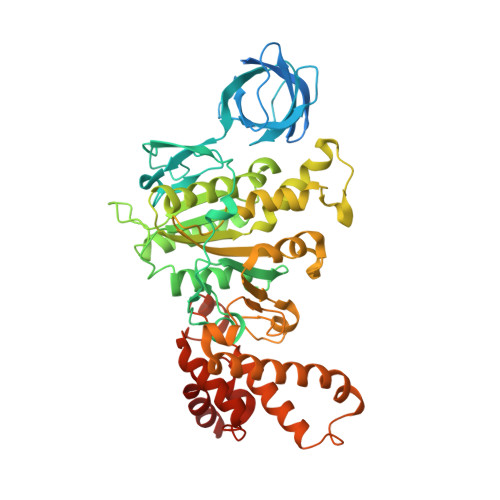
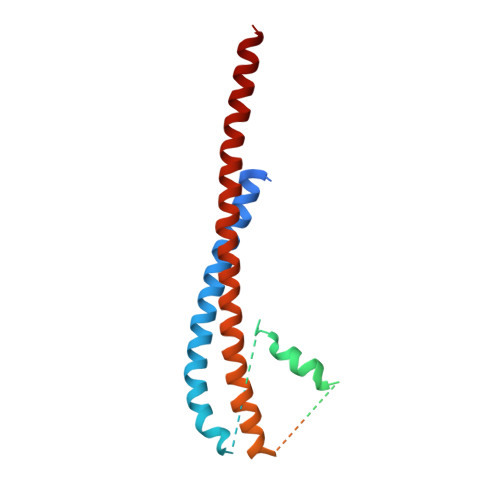
Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Sanchez-Weatherby, J., Bowler, M.W., Huet, J., Gobbo, A., Felisaz, F., Lavault, B., Moya, R., Kadlec, J., Ravelli, R.B., Cipriani, F.(2009) Acta Crystallogr D Biol Crystallogr 65: 1237-1246
- PubMed: 19966409
- DOI: https://doi.org/10.1107/S0907444909037822
- Primary Citation of Related Structures:
2W6E, 2W6F, 2W6G, 2W6H, 2W6I, 2W6J - PubMed Abstract:
Dehydration of protein crystals is rarely used, despite being a post-crystallization method that is useful for the improvement of crystal diffraction properties, as it is difficult to reproduce and monitor. A novel device for hydration control of macromolecular crystals in a standard data-collection environment has been developed. The device delivers an air stream of precise relative humidity that can be used to alter the amount of water in macromolecular crystals. The device can be rapidly installed and is fully compatible with most standard synchrotron X-ray beamlines. Samples are mounted in cryoloops and the progress of dehydration can be monitored both optically and by the acquisition of diffraction images. Once the optimal hydration level has been obtained, cryocooling is easy to achieve by hand or by using a sample changer. The device has been thoroughly tested on several ESRF beamlines and is available to users.
- Diamond Light Source Ltd, Diamond House DR1.40, Harwell Science and Innovation Campus, RAL, Chilton, Didcot, Oxfordshire OX11 0DE, England. juan.sanchez-weatherby@diamond.ac.uk
Organizational Affiliation: