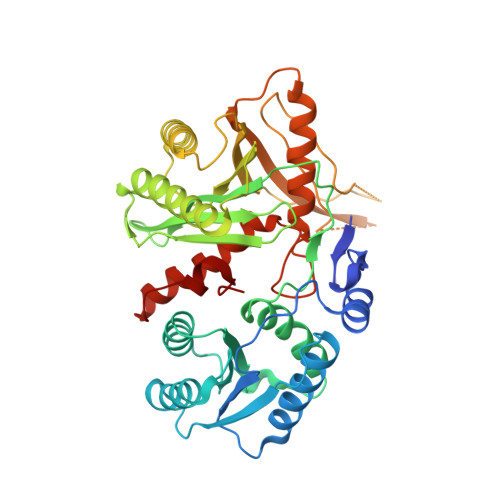
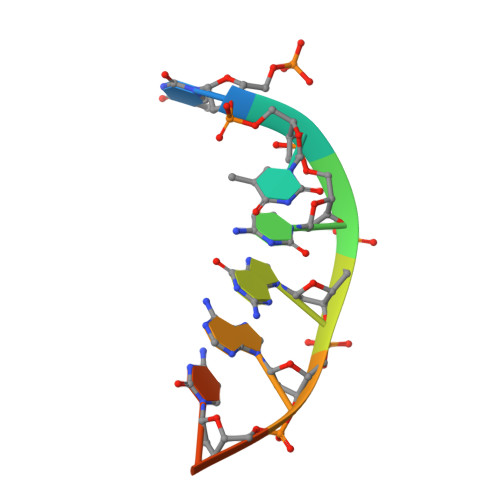
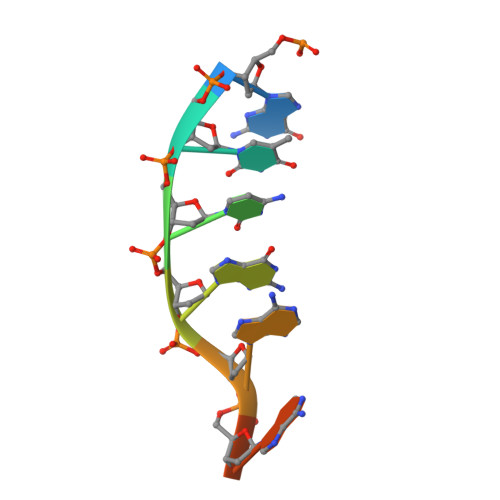
Enhancement of the Seed-Target Recognition Step in RNA Silencing by a Piwi-Mid Domain Protein
Parker, J.S., Parizotto, E.A., Wang, M., Roe, S.M., Barford, D.(2009) Mol Cell 33: 204
- PubMed: 19187762
- DOI: https://doi.org/10.1016/j.molcel.2008.12.012
- Primary Citation of Related Structures:
2W42 - PubMed Abstract:
Target recognition in RNA silencing is governed by the "seed sequence" of a guide RNA strand associated with the PIWI/MID domain of an Argonaute protein in RISC. Using a reconstituted in vitro target recognition system, we show that a model PIWI/MID domain protein confers position-dependent tightening and loosening of guide-strand-target interactions. Over the seed sequence, the interaction affinity is enhanced up to approximately 300-fold. Enhancement is achieved through a reduced entropy penalty for the interaction. In contrast, interactions 3' of the seed are inhibited. We quantified mismatched target recognition inside and outside the seed, revealing amplified discrimination at the third position in the seed mediated by the PIWI/MID domain. Thus, association of the guide strand with the PIWI/MID domain generates an enhanced affinity anchor site over the seed that can promote fidelity in target recognition and stabilize and guide the assembly of the active silencing complex.
- Institute of Cancer Research, Chester Beatty Laboratories, London, UK. james.parker@bioch.ox.ac.uk
Organizational Affiliation: