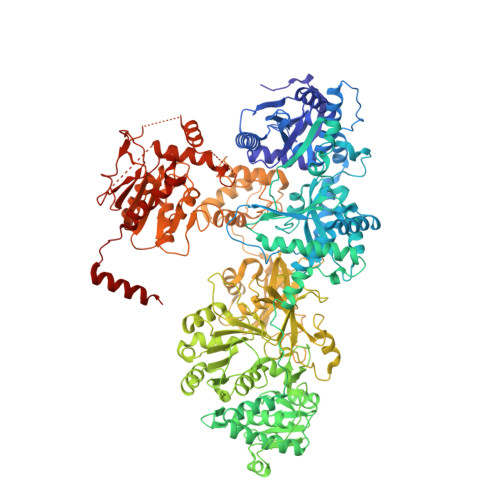
Crystal structure of the termination module of a nonribosomal peptide synthetase.
Tanovic, A., Samel, S.A., Essen, L.O., Marahiel, M.A.(2008) Science 321: 659-663
- PubMed: 18583577
- DOI: https://doi.org/10.1126/science.1159850
- Primary Citation of Related Structures:
2VSQ - PubMed Abstract:
Nonribosomal peptide synthetases (NRPSs) are modular multidomain enzymes that act as an assembly line to catalyze the biosynthesis of complex natural products. The crystal structure of the 144-kilodalton Bacillus subtilis termination module SrfA-C was solved at 2.6 angstrom resolution. The adenylation and condensation domains of SrfA-C associate closely to form a catalytic platform, with their active sites on the same side of the platform. The peptidyl carrier protein domain is flexibly tethered to this platform and thus can move with its substrate-loaded 4'-phosphopantetheine arm between the active site of the adenylation domain and the donor side of the condensation domain. The SrfA-C crystal structure has implications for the rational redesign of NRPSs as a means of producing novel bioactive peptides.
- Biochemistry, Department of Chemistry, Philipps University Marburg, Hans-Meerwein-Strasse, D35032 Marburg, Germany.
Organizational Affiliation: