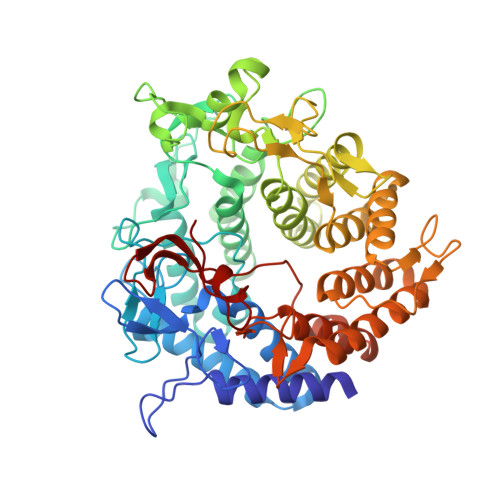
A Family 2 Pectate Lyase Displays a Rare Fold and Transition Metal-Assisted -Elimination.
Abbott, D.W., Boraston, A.B.(2007) J Biological Chem 282: 35328
- PubMed: 17881361
- DOI: https://doi.org/10.1074/jbc.M705511200
- Primary Citation of Related Structures:
2V8I, 2V8J, 2V8K - PubMed Abstract:
The family 2 pectate lyase from Yersinia enterocolitica (YePL2A), solved to 1.5A, reveals it to be the first prokaryotic protein reported to display the rare (alpha/alpha)(7) barrel fold. In addition to its apo form, we have also determined the structure of a metal-bound form of YePL2A (to 2.0A) and a trigalacturonic acid-bound substrate complex (to 2.1A) Although its fold is rare, the catalytic center of YePL2A can be superimposed with structurally unrelated families, underlining the conserved catalytic amino acid architecture of the beta-elimination mechanism. In addition to its overall structure, YePL2A also has two other unique features: 1) it utilizes a metal atom other than calcium for catalysis, and 2) its Brønstead base is in an alternate conformation and directly interacts with the uronate group of the substrate.
- Biochemistry & Microbiology, University of Victoria, Victoria, British Columbia V8W 3P6, Canada.
Organizational Affiliation: