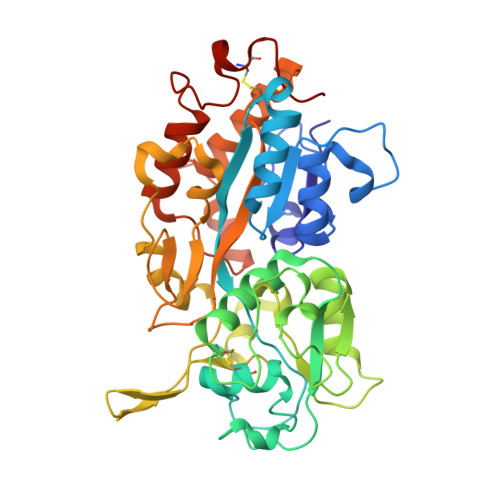
Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Diemer, H., Elias, M., Renault, F., Rochu, D., Contreras-Martel, C., Schaeffer, C., Van Dorsselaer, A., Chabriere, E.(2008) Proteins 71: 1708
- PubMed: 18076037
- DOI: https://doi.org/10.1002/prot.21866
- Primary Citation of Related Structures:
2V3Q - PubMed Abstract:
The Human Phosphate Binding Protein (HPBP) is a serendipitously discovered apolipoprotein from human plasma that binds phosphate. Amino acid sequence relates HPBP to an intriguing protein family that seems ubiquitous in eukaryotes. These proteins, named DING according to the sequence of their four conserved N-terminal residues, are systematically absent from eukaryotic genome databases. As a consequence, HPBP amino acids sequence had to be first assigned from the electronic density map. Then, an original approach combining X-ray crystallography and mass spectrometry provides the complete and a priori exact sequence of the 38-kDa HPBP. This first complete sequence of a eukaryotic DING protein will be helpful to study HPBP and the entire DING protein family.
- Laboratoire de Spectrométrie de Masse Bioorganique, Institut Pluridisciplinaire Hubert Curien, UMR 7178 (CNRS-ULP) ECPM, 25 rue Becquerel F67087-Strasbourg-Cedex 2, France.
Organizational Affiliation: