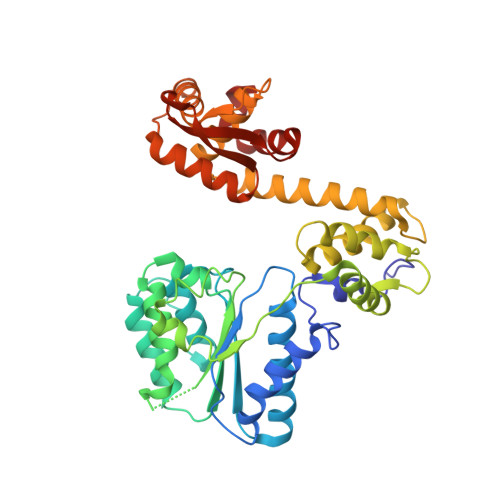
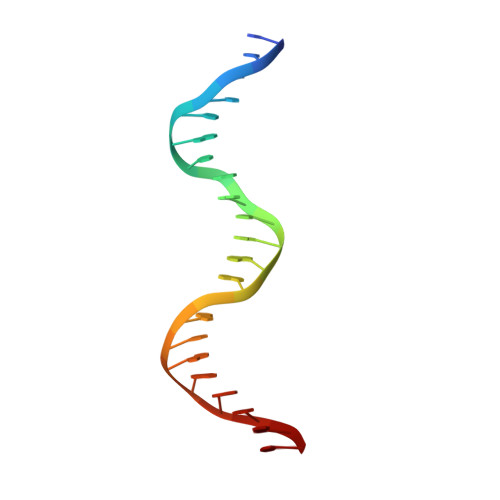
Structural Basis of DNA Replication Origin Recognition by an Orc Protein.
Gaudier, M., Schuwirth, B.S., Westcott, S.L., Wigley, D.B.(2007) Science 317: 1213
- PubMed: 17761880
- DOI: https://doi.org/10.1126/science.1143664
- Primary Citation of Related Structures:
2V1U - PubMed Abstract:
DNA replication in archaea and in eukaryotes share many similarities. We report the structure of an archaeal origin recognition complex protein, ORC1, bound to an origin recognition box, a DNA sequence that is found in multiple copies at replication origins. DNA binding is mediated principally by a C-terminal winged helix domain that inserts deeply into the major and minor grooves, widening them both. However, additional DNA contacts are made with the N-terminal AAA+ domain, which inserts into the minor groove at a characteristic G-rich sequence, inducing a 35 degrees bend in the duplex and providing directionality to the binding site. Both contact regions also induce substantial unwinding of the DNA. The structure provides insight into the initial step in assembly of a replication origin and recruitment of minichromosome maintenance (MCM) helicase to that origin.
- Cancer Research UK Clare Hall Laboratories, London Research Institute, Blanche Lane, South Mimms, Potters Bar, Herts EN6 3LD, UK.
Organizational Affiliation: