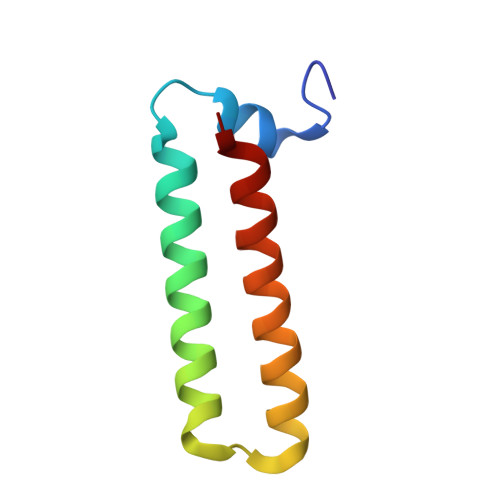
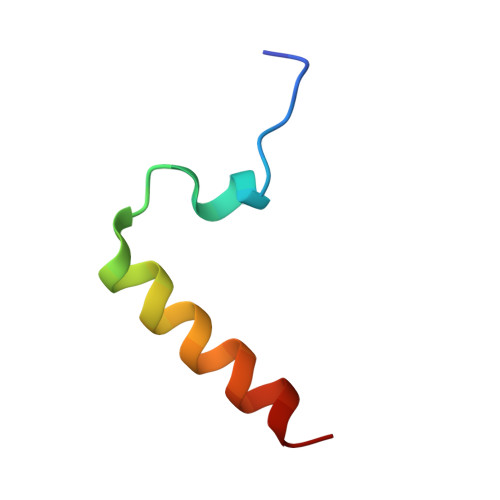
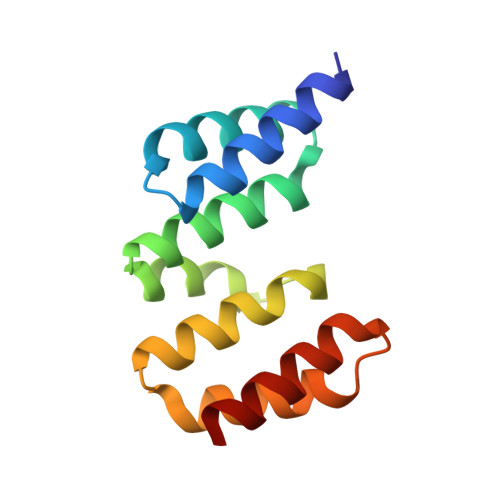
Structure of the heterotrimeric complex that regulates type III secretion needle formation.
Quinaud, M., Ple, S., Job, V., Contreras-Martel, C., Simorre, J.P., Attree, I., Dessen, A.(2007) Proc Natl Acad Sci U S A 104: 7803-7808
- PubMed: 17470796
- DOI: https://doi.org/10.1073/pnas.0610098104
- Primary Citation of Related Structures:
2UWJ - PubMed Abstract:
Type III secretion systems (T3SS), found in several Gram-negative pathogens, are nanomachines involved in the transport of virulence effectors directly into the cytoplasm of target cells. T3SS are essentially composed of basal membrane-embedded ring-like structures and a hollow needle formed by a single polymerized protein. Within the bacterial cytoplasm, the T3SS needle protein requires two distinct chaperones for stabilization before its secretion, without which the entire T3SS is nonfunctional. The 2.0-A x-ray crystal structure of the PscE-PscF(55-85)-PscG heterotrimeric complex from Pseudomonas aeruginosa reveals that the C terminus of the needle protein PscF is engulfed within the hydrophobic groove of the tetratricopeptide-like molecule PscG, indicating that the macromolecular scaffold necessary to stabilize the T3SS needle is totally distinct from chaperoned complexes between pilus- or flagellum-forming molecules. Disruption of specific PscG-PscF interactions leads to impairment of bacterial cytotoxicity toward macrophages, indicating that this essential heterotrimer, which possesses homologs in a wide variety of pathogens, is a unique attractive target for the development of novel antibacterials.
- Institut de Biologie Structurale Jean-Pierre Ebel, 41 Rue Jules Horowitz, Unité Mixte de Recherche 5075, Commissariat à l'Energie Atomique, Centre National de la Recherche Scientifique, Université Joseph Fourier, 38027 Grenoble, France.
Organizational Affiliation: