Study of the structure and dynamics of a chimeric variant of the SH3 domain (SHA-Bergerac) by NMR spectroscopy
Prokhorov, D.A., Timchenko, M.A., Kudrevatykh, Y.A., Fedyukina, D.V., Gushchina, L.V., Khristoforov, V.S., Filimonov, V.V., Kutyshenko, V.P.(2008) Russ J Bioorg Chem 34: 578-585
- PubMed: 19060939
- DOI: https://doi.org/10.1134/s1068162008050075
- Primary Citation of Related Structures:
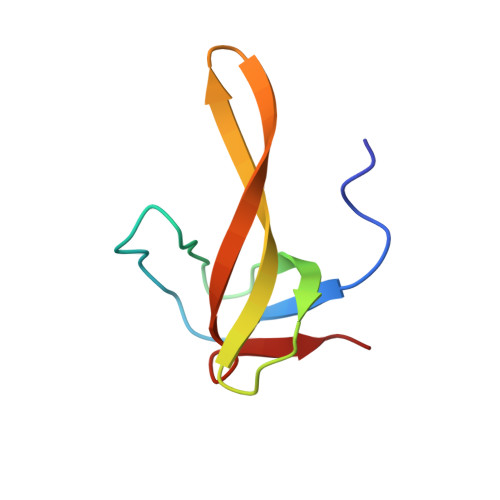
2RMO - PubMed Abstract:
A structural-dynamic study of one of the chimeric proteins (SHA) belonging to the SH3-Bergerac family and containing the KATANGKTYE sequence instead of the N47D48 beta-turn in the spectrin SH3 domain was carried out by high resolution NMR spectroscopy. The spatial structure of the protein was determined and its dynamics in solution was investigated on the basis of the NMR data. The elongation of the SHA polypeptide chain in comparison with the WT-SH3 original protein (by ~17%) exerts practically no effect on the general topology of the molecule. The presence of a stable beta-hairpin in the region of insertion was confirmed. This hairpin was shown to have a higher mobility in comparison with other regions of the protein.